Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate
- PMID: 9738025
- PMCID: PMC105069
- DOI: 10.1128/JCM.36.10.2810-2816.1998
Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate
Abstract
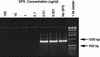
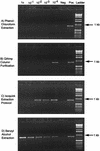
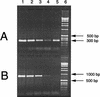
Molecular methods are increasingly used to identify microbes in clinical samples. A common technical problem with PCR is failed amplification due to the presence of PCR inhibitors. Initial attempts at amplification of the bacterial 16S rRNA gene from inoculated blood culture media failed for this reason. The inhibitor persisted, despite numerous attempts to purify the DNA, and was identified as sodium polyanetholesulfonate (SPS), a common additive to blood culture media. Like DNA, SPS is a high-molecular-weight polyanion that is soluble in water but insoluble in alcohol. Accordingly, SPS tends to copurify with DNA. An extraction method was designed for purification of DNA from blood culture media and removal of SPS. Blood culture media containing human blood and spiked with Escherichia coli was subjected to an organic extraction procedure with benzyl alcohol, and removal of SPS was documented spectrophotometrically. Successful amplification of the extracted E. coli 16S rRNA gene was achieved by adding 5 microliter of undiluted processed sample DNA to a 50-microliter PCR mixture. When using other purification methods, the inhibitory effect of SPS could be overcome only by dilution of these samples. By our extraction technique, even uninoculated blood culture media were found to contain bacterial DNA when they were subjected to broad-range 16S rRNA gene consensus PCR. We conclude that the blood culture additive SPS is a potent inhibitor of PCR, is resistant to removal by traditional DNA purification methods, but can be removed by a benzyl alcohol extraction protocol that results in improved PCR performance.
Figures



References
-
- Ahokas H, Erkkila M J. Interference of PCR amplification by the polyamines, spermine and spermidine. PCR Methods Appl. 1993;3:65–68. - PubMed
-
- Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J Forensic Sci. 1994;39:362–372. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

