Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses
- PMID: 9738055
- PMCID: PMC105099
- DOI: 10.1128/JCM.36.10.2990-2995.1998
Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses
Abstract
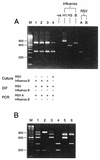
A multiplex reverse transcription (RT)-PCR method that has been developed is capable of detecting and subtyping influenza A (H1N1 and H3N2) and B viruses as well as respiratory syncytial virus (RSV) types A and B in respiratory clinical samples taken as part of a national community-based surveillance program of influenza-like illness in England and Wales. The detection of each different pathogen depended on distinguishing five amplification products of different sizes on agarose gels following RT-PCR with multiple primer sets. The multiplex RT-PCR was tested with 65 nasopharyngeal apirates from which RSV had been isolated and 237 combined nose and throat swabs from which influenza A (H1N1 and H3N2) or B virus had been detected by virus isolation, as well as 40 respiratory samples from which other viruses including cytomegalovirus, herpes simplex virus, enteroviruses, and parainfluenza viruses had been grown. For the typing and subtyping of influenza A and B viruses and RSV types A and B, the multiplex RT-PCR gave an excellent (100%) correlation with the results of conventional typing and subtyping with specific antisera. Multiplex RT-PCR can also be used to accurately detect more than one viral template in the same reaction mixture, allowing viral coinfections to be identified with the same respiratory specimen.
Figures




References
-
- Anderson L J, Hierholzer J C, Tsou C, Hendry R M, Fernie B F, Stone Y, McIntosh K. Antigenic characterisation of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis. 1985;163:626–633. - PubMed
-
- Axton R A, Brock D J. A single tube multiplex system for the simultaneous detection of 10 common cystic fibrosis mutations. Hum Mutat. 1995;5:260–262. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

