Degradation of myogenic transcription factor MyoD by the ubiquitin pathway in vivo and in vitro: regulation by specific DNA binding
- PMID: 9742084
- PMCID: PMC109153
- DOI: 10.1128/MCB.18.10.5670
Degradation of myogenic transcription factor MyoD by the ubiquitin pathway in vivo and in vitro: regulation by specific DNA binding
Abstract
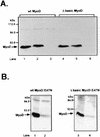
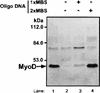
MyoD is a tissue-specific transcriptional activator that acts as a master switch for skeletal muscle differentiation. Its activity is induced during the transition from proliferating, nondifferentiated myoblasts to resting, well-differentiated myotubes. Like many other transcriptional regulators, it is a short-lived protein; however, the targeting proteolytic pathway and the underlying regulatory mechanisms involved in the process have remained obscure. It has recently been shown that many short-lived regulatory proteins are degraded by the ubiquitin system. Degradation of a protein by the ubiquitin system proceeds via two distinct and successive steps, conjugation of multiple molecules of ubiquitin to the target protein and degradation of the tagged substrate by the 26S proteasome. Here we show that MyoD is degraded by the ubiquitin system both in vivo and in vitro. In intact cells, the degradation is inhibited by lactacystin, a specific inhibitor of the 26S proteasome. Inhibition is accompanied by accumulation of high-molecular-mass MyoD-ubiquitin conjugates. In a cell-free system, the proteolytic process requires both ATP and ubiquitin and, like the in vivo process, is preceded by formation of ubiquitin conjugates of the transcription factor. Interestingly, the process is inhibited by the specific DNA sequence to which MyoD binds: conjugation and degradation of a MyoD mutant protein which lacks the DNA-binding domain are not inhibited. The inhibitory effect of the DNA requires the formation of a complex between the DNA and the MyoD protein. Id1, which inhibits the binding of MyoD complexes to DNA, abrogates the effect of DNA on stabilization of the protein.
Figures

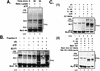




References
-
- Bates S, Vousden K H. p53 in signaling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6:1–7. - PubMed
-
- Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. - PubMed
-
- Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz A L, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem. 1997;272:9002–9010. - PubMed
-
- Blumenfeld N, Gonen H, Mayer A, Smith C E, Siegel N R, Schwartz A L, Ciechanover A. Purification and characterization of a novel species of ubiquitin-carrier protein, E2, that is involved in degradation of non-“N-end rule” protein substrates. J Biol Chem. 1994;269:9574–9581. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
