Influence of intron length on alternative splicing of CD44
- PMID: 9742110
- PMCID: PMC109179
- DOI: 10.1128/MCB.18.10.5930
Influence of intron length on alternative splicing of CD44
Abstract
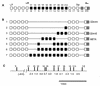
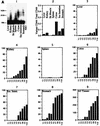
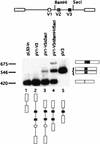
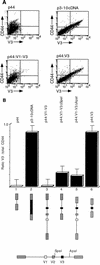
Although the splicing of transcripts from most eukaryotic genes occurs in a constitutive fashion, some genes can undergo a process of alternative splicing. This is a genetically economical process which allows a single gene to give rise to several protein isoforms by the inclusion or exclusion of sequences into or from the mature mRNA. CD44 provides a unique example; more than 1,000 possible isoforms can be produced by the inclusion or exclusion of a central tandem array of 10 alternatively spliced exons. Certain alternatively spliced exons have been ascribed specific functions; however, independent regulation of the inclusion or skipping of each of these exons would clearly demand an extremely complex regulatory network. Such a network would involve the interaction of many exon-specific trans-acting factors with the pre-mRNA. Therefore, to assess whether the exons are indeed independently regulated, we have examined the alternative exon content of a large number of individual CD44 cDNA isoforms. This analysis shows that the downstream alternatively spliced exons are favored over those lying upstream and that alternative exons are often included in blocks rather than singly. Using a novel in vivo alternative splicing assay, we show that intron length has a major influence upon the alternative splicing of CD44. We propose a kinetic model in which short introns may overcome the poor recognition of alternatively spliced exons. These observations suggest that for CD44, intron length has been exploited in the evolution of the genomic structure to enable tissue-specific patterns of splicing to be maintained.
Figures


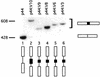
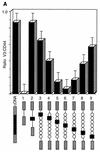


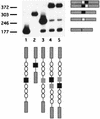
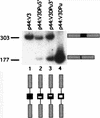
Similar articles
-
The architecture of pre-mRNAs affects mechanisms of splice-site pairing.Proc Natl Acad Sci U S A. 2005 Nov 8;102(45):16176-81. doi: 10.1073/pnas.0508489102. Epub 2005 Oct 31. Proc Natl Acad Sci U S A. 2005. PMID: 16260721 Free PMC article.
-
Gene therapeutic approach to primary and metastatic brain tumors: I. CD44 variant pre-RNA alternative splicing as a CEPT control element.J Neurooncol. 1995 Dec;26(3):243-50. doi: 10.1007/BF01052627. J Neurooncol. 1995. PMID: 8750190
-
Alternative spliced transcripts as cancer markers.Dis Markers. 2001;17(2):67-75. doi: 10.1155/2001/184856. Dis Markers. 2001. PMID: 11673653 Free PMC article. Review.
-
Multisite and bidirectional exonic splicing enhancer in CD44 alternative exon v3.RNA. 2007 Dec;13(12):2312-23. doi: 10.1261/rna.732807. Epub 2007 Oct 16. RNA. 2007. PMID: 17940137 Free PMC article.
-
[Molecular mechanism of mRNA alternative splicing].Yi Chuan Xue Bao. 2004 Jan;31(1):102-7. Yi Chuan Xue Bao. 2004. PMID: 15468927 Review. Chinese.
Cited by
-
Two classes of genes in plants.Genetics. 2000 Apr;154(4):1819-25. doi: 10.1093/genetics/154.4.1819. Genetics. 2000. PMID: 10747072 Free PMC article.
-
The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4.EMBO J. 2001 Jul 16;20(14):3821-30. doi: 10.1093/emboj/20.14.3821. EMBO J. 2001. PMID: 11447123 Free PMC article.
-
NSrp70 suppresses metastasis in triple-negative breast cancer by modulating Numb/TβR1/EMT axis.Oncogene. 2022 Jun;41(25):3409-3422. doi: 10.1038/s41388-022-02349-z. Epub 2022 May 14. Oncogene. 2022. PMID: 35568738
-
Unique CD44 intronic SNP is associated with tumor grade in breast cancer: a case control study and in silico analysis.Cancer Cell Int. 2018 Feb 23;18:28. doi: 10.1186/s12935-018-0522-2. eCollection 2018. Cancer Cell Int. 2018. PMID: 29483847 Free PMC article.
-
Splicing of long non-coding RNAs primarily depends on polypyrimidine tract and 5' splice-site sequences due to weak interactions with SR proteins.Nucleic Acids Res. 2019 Jan 25;47(2):911-928. doi: 10.1093/nar/gky1147. Nucleic Acids Res. 2019. PMID: 30445574 Free PMC article.
References
-
- Arch R, Wirth K, Hofmann M, Ponta H, Matzku S, Herrlich P, Zoller M. Participation in normal immune responses of a metastasis-inducing splice variant of CD44. Science. 1992;257:682–685. - PubMed
-
- Aruffo A, Stamenkovic I, Melnick M, Underhill C B, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–1313. - PubMed
-
- Bartolazzi A, Jackson D, Bennett K, Aruffo A, Dickinson R, Shields J, Whittle N, Stamenkovic I. Regulation of growth and dissemination of a human lymphoma by CD44 splice variants. J Cell Sci. 1995;108:1723–1733. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
