Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators
- PMID: 9742131
- PMCID: PMC109200
- DOI: 10.1128/MCB.18.10.6142
Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators
Abstract
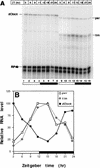
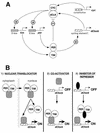
The Clock gene plays an essential role in the manifestation of circadian rhythms (approximately 24 h) in mice and is a member of the basic helix-loop-helix (bHLH) PER-ARNT-SIM (PAS) superfamily of transcription factors. Here we report the characterization of a novel Drosophila bHLH-PAS protein that is highly homologous to mammalian CLOCK. (Similar findings were recently described by Allada et al. Cell 93:791-804, 1998, and Darlington et al., Science 280:1599-1603, 1998.) Transcripts from this putative Clock ortholog (designated dClock) undergo daily rhythms in abundance that are antiphase to the cycling observed for the RNA products from the Drosophila melanogaster circadian clock genes period (per) and timeless (tim). Furthermore, dClock RNA cycling is abolished and the levels are at trough values in the absence of either PER or TIM, suggesting that these two proteins can function as transcriptional activators, a possibility which is in stark contrast to their previously characterized role in transcriptional autoinhibition. Finally, the temporal regulation of dClock expression is quickly perturbed by shifts in light-dark cycles, indicating that this molecular rhythm is closely connected to the photic entrainment pathway. The isolation of a Drosophila homolog of Clock together with the recent discovery of mammalian homologs of per indicate that there is high structural conservation in the integral components underlying circadian oscillators in Drosophila and mammals. Nevertheless, because mammalian Clock mRNA is constitutively expressed, our findings are a further example of striking differences in the regulation of putative circadian clock orthologs in different species.
Figures

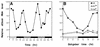

References
-
- Albrecht U, Sun Z S, Eichele G, Lee C C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. - PubMed
-
- Allada R, White N E, So W V, Hall J C, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. - PubMed
-
- Aronson B D, Johnson K A, Loros J J, Dunlap J C. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. - PubMed
-
- Citri Y, Colot H V, Jacquier A C, Yu Q, Hall J C, Baltimore D, Rosbash M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature. 1987;326:42–47. - PubMed
-
- Crews S T. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998;12:607–620. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
