Molecular and biological characterization of the murine leukotriene B4 receptor expressed on eosinophils
- PMID: 9743525
- PMCID: PMC2212531
- DOI: 10.1084/jem.188.6.1063
Molecular and biological characterization of the murine leukotriene B4 receptor expressed on eosinophils
Abstract
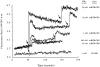
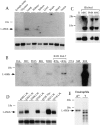
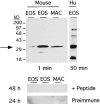
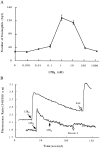
The movement of leukocytes into tissues is regulated by the local production of chemical mediators collectively referred to as chemoattractants. Although chemoattractants constitute a diverse array of molecules, including proteins, peptides, and lipids, they all appear to signal leukocytes through a related family of seven transmembrane-spanning G protein-coupled receptors. The eosinophil is a potent proinflammatory cell that is attracted into tissues during allergic inflammation, parasitic infection, and certain malignancies. Since the molecular mechanisms controlling eosinophil recruitment are incompletely understood, we performed a degenerate polymerase chain reaction on cDNA isolated from murine eosinophils to identify novel chemoattractant receptors. We report the isolation of a cDNA that encodes a 351-amino acid glycoprotein that is 78% identical to a human gene that has been reported to be a purinoceptor (P2Y7) and a leukotriene B4 (LTB4) receptor (BLTR). Chinese hamster ovary (CHO) cells transfected with this cDNA specifically bound [3H]LTB4 with a dissociation constant of 0.6 +/- 0.1 nM. Furthermore, LTB4 induced a dose-dependent intracellular calcium flux in transfected CHO cells. In contrast, [35S]dATP did not specifically bind to these transfectants. This mRNA was expressed at high levels in interleukin 5-exposed eosinophils, elicited peritoneal macrophages and neutrophils, and to a lesser extent interferon gamma stimulated macrophages. Low levels of expression were detected in the lung, lymph node, and spleen of unchallenged mice. Western blot analysis detected the mBLTR protein in murine eosinophils and alveolar macrophages as well as human eosinophils. In addition, elevated levels of mBLTR mRNA were found in the lungs of mice in a murine model of allergic pulmonary inflammation in a time course consistent with the influx of eosinophils. Our findings indicate that this murine receptor is an LTB4 receptor that is highly expressed on activated leukocytes, including eosinophils, and may play an important role in mediating eosinophil recruitment into inflammatory foci.
Figures






References
-
- Weller PF. The immunobiology of eosinophils. N Eng J Med. 1991;324:1110–1118. - PubMed
-
- Bousquett J, Chanez P, Lacoste JY, Barneon G, Ghavanian N, Enander I, Venge P, Ahlstedt S, Simony-Lafontaine J, et al. Eosinophilic inflammation in asthma. N Eng J Med. 1990;323:1033–1039. - PubMed
-
- Bochner BS, Schleimer RP. The role of adhesion molecules in human eosinophil and basophil recruitment. J Allergy Clin Immunol. 1994;94:427–428. - PubMed
-
- Resnick MB, Weller PF. Mechanisms of eosinophil recruitment. Am J Respir Cell Mol Biol. 1993;8:349–355. - PubMed
-
- Baggiolini M, Dahinden CA. CC chemokines in allergic inflammation. Immunol Today. 1994;15:127–133. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

