Major histocompatibility complex class I viral antigen processing in the secretory pathway defined by the trans-Golgi network protease furin
- PMID: 9743529
- PMCID: PMC2212533
- DOI: 10.1084/jem.188.6.1105
Major histocompatibility complex class I viral antigen processing in the secretory pathway defined by the trans-Golgi network protease furin
Abstract
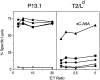
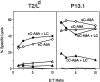
Classical antigen presentation by major histocompatibility complex class I molecules involves cytosolic processing of endogenously synthesized antigens by proteasomes and translocation of processed peptides into the endoplasmic reticulum (ER) by transporters associated with antigen presentation (TAP). Alternative pathways for processing of endogenous antigens, generally involving the ER, have been suggested but not fully proved. We analyzed the potential for class I presentation of proteolytic maturation of secretory antigens in the exocytic pathway. We found that hepatitis B (HB) virus secretory core protein HBe can efficiently deliver COOH-terminally located antigenic peptides for endogenous class I loading in the absence of TAP. Antigen presentation to specific cytotoxic T lymphocytes correlates with protein maturation at the COOH terminus, since modification of maturation and transport of HBe through the secretory pathway alters antigen presentation. Both maturation and a necessary processing step occur in the Golgi or post-Golgi compartment. Antigen presentation is independent of proteasome activity, but inhibitors of the trans-Golgi network resident protease furin inhibit both HBe maturation and antigen presentation. These results define a new antigen processing pathway located in the secretory route, with a central role for proteolytic maturation mediated by the subtilisin protease family member furin as an efficient source for antigen presentation.
Figures
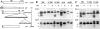




References
-
- Matsumura M, Fremont DH, Peterson PA, Wilson IA. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992;257:927–934. - PubMed
-
- Bouvier M, Wiley DC. Importance of peptide amino and carboxyl termini to the stability of MHC class I molecules. Science. 1994;265:398–402. - PubMed
-
- York IA, Rock KL. Antigen processing and presentation by the class I major histocompatibility complex. Annu Rev Immunol. 1996;14:369–396. - PubMed
-
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasome. Annu Rev Biochem. 1996;65:801–847. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

