Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway
- PMID: 9744871
- PMCID: PMC317167
- DOI: 10.1101/gad.12.18.2956
Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway
Abstract
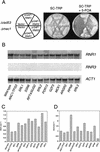
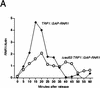
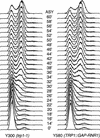
RAD53 and MEC1 are essential genes required for the transcriptional and cell cycle responses to DNA damage and DNA replication blocks. We have examined the essential function of these genes and found that their lethality but not their checkpoint defects can be suppressed by increased expression of genes encoding ribonucleotide reductase. Analysis of viable null alleles revealed that Mec1 plays a greater role in response to inhibition of DNA synthesis than Rad53. The loss of survival in mec1 and rad53 null or point mutants in response to transient inhibition of DNA synthesis is not a result of inappropriate anaphase entry but primarily to an inability to complete chromosome replication. We propose that this checkpoint pathway plays an important role in the maintenance of DNA synthetic capabilities when DNA replication is stressed.
Figures











References
-
- Allen JB, Elledge SJ. A family of vectors that facilitate transposon and insertional mutagenesis of cloned genes in yeast. Yeast. 1994;10:1267–1272. - PubMed
-
- Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes & Dev. 1994;8:2416–2428. - PubMed
-
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, Wynshaw-Boris A. Atm-deficient mice: A paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. - PubMed
-
- Carr AM, Moudjou M, Bentley NJ, Hagan IM. The chk1 pathway is required to prevent mitosis following cell-cycle arrest at ‘start’. Curr Biol. 1995;5:1179–1190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
