The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge
- PMID: 9744887
- PMCID: PMC2141774
- DOI: 10.1083/jcb.142.6.1595
The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge
Abstract
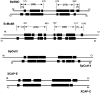
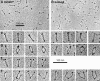
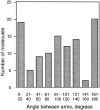
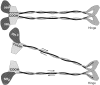
Structural maintenance of chromosomes (SMC) proteins function in chromosome condensation and several other aspects of DNA processing. They are large proteins characterized by an NH2-terminal nucleotide triphosphate (NTP)-binding domain, two long segments of coiled coil separated by a hinge, and a COOH-terminal domain. Here, we have visualized by EM the SMC protein from Bacillus subtilis (BsSMC) and MukB from Escherichia coli, which we argue is a divergent SMC protein. Both BsSMC and MukB show two thin rods with globular domains at the ends emerging from the hinge. The hinge appears to be quite flexible: the arms can open up to 180 degrees, separating the terminal domains by 100 nm, or close to near 0 degrees, bringing the terminal globular domains together. A surprising observation is that the approximately 300-amino acid-long coiled coils are in an antiparallel arrangement. Known coiled coils are almost all parallel, and the longest antiparallel coiled coils known previously are 35-45 amino acids long. This antiparallel arrangement produces a symmetrical molecule with both an NH2- and a COOH-terminal domain at each end. The SMC molecule therefore has two complete and identical functional domains at the ends of the long arms. The bifunctional symmetry and a possible scissoring action at the hinge should provide unique biomechanical properties to the SMC proteins.
Figures




References
-
- Abrahams JP. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. - PubMed
-
- Erickson, H.P., and G. Briscoe. 1995. Tenascin, laminin and fibronectin produced by cultured cells. In Extracellular Matrix: A Practical Approach. M.A. Haralson and J.R. Hassell, editors. Oxford University Press, Oxford, UK. 187–198.
-
- Fowler WE, Erickson HP. Trinodular structure of fibrinogen. Confirmation by both shadowing and negative stain electron microscopy. J Mol Biol. 1979;134:241–249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

