Isolation of Candida glabrata homologs of the Saccharomyces cerevisiae KRE9 and KNH1 genes and their involvement in cell wall beta-1,6-glucan synthesis
- PMID: 9748432
- PMCID: PMC107535
- DOI: 10.1128/JB.180.19.5020-5029.1998
Isolation of Candida glabrata homologs of the Saccharomyces cerevisiae KRE9 and KNH1 genes and their involvement in cell wall beta-1,6-glucan synthesis
Abstract
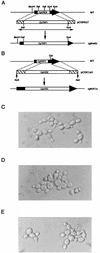
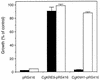
The Candida glabrata KRE9 (CgKRE9) and KNH1 (CgKNH1) genes have been isolated as multicopy suppressors of the tetracycline-sensitive growth of a Saccharomyces cerevisiae mutant with the disrupted KNH1 locus and the KRE9 gene placed under the control of a tetracycline-responsive promoter. Although a cgknh1Delta mutant showed no phenotype beyond slightly increased sensitivity to the K1 killer toxin, disruption of CgKRE9 resulted in several phenotypes similar to those of the S. cerevisiae kre9Delta null mutant: a severe growth defect on glucose medium, resistance to the K1 killer toxin, a 50% reduction of beta-1,6-glucan, and the presence of aggregates of cells with abnormal morphology on glucose medium. Replacement in C. glabrata of the cognate CgKRE9 promoter with the tetracycline-responsive promoter in a cgknh1Delta background rendered cell growth tetracycline sensitive on media containing glucose or galactose. cgkre9Delta cells were shown to be sensitive to calcofluor white specifically on glucose medium. In cgkre9 mutants grown on glucose medium, cellular chitin levels were massively increased.
Figures





References
-
- Aisner J, Schimpff S C, Sutherland J C, Young V M, Wiernik P H. Torulopsis glabrata infections in patients with cancer: increasing incidence and relationship to colonization. Am J Med. 1976;61:23–28. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

