New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating
- PMID: 9748433
- PMCID: PMC107536
- DOI: 10.1128/JB.180.19.5030-5037.1998
New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating
Abstract
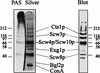
Biotinylation of intact Saccharomyces cerevisiae cells with a nonpermeant reagent (Sulfo-NHS-LC-Biotin) allowed the identification of seven cell wall proteins that were released from intact cells by dithiothreitol (DTT). By N-terminal sequencing, three of these proteins were identified as the known proteins beta-exoglucanase 1 (Exg1p), beta-endoglucanase (Bgl2p), and chitinase (Cts1p). One protein was related to the PIR protein family, whereas the remaining three (Scw3p, Scw4p, and Scw10p [for soluble cell wall proteins]) were found to be related to glucanases. Single knockouts of these three potential glucanases did not result in dramatic phenotypes. The double knockout of SCW4 and the homologous gene SCW10 resulted in slower growth, significantly increased release of proteins from intact cells by DTT, and highly decreased mating efficiency when these two genes were disrupted in both mating types. The synergistic behavior of the disruption of SCW4 and SCW10 was partly antagonized by the disruption of BGL2. The data are discussed in terms of a possible counterplay of transglucosidase and glucosidase activities.
Figures








References
-
- Angiolella L, Facchin M, Stringaro A, Maras B, Simonetti N, Cassone A. Identification of a glucan-associated enolase as a main cell wall protein of Candida albicansand an indirect target of lipopeptide antimycotics. J Infect Dis. 1996;173:684–690. - PubMed
-
- Broach J R, Jones E W, Pringle J R, editors. The molecular and cellular biology of the yeast Saccharomyces. 1. Genome dynamics, protein synthesis and energetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1991.
-
- Cappellaro, C. Unpublished data.
-
- Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, et al. Life with 6000 genes. Science. 1996;274:546–567. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

