Filamentous bacteriophages of Vibrio parahaemolyticus as a possible clue to genetic transmission
- PMID: 9748441
- PMCID: PMC107544
- DOI: 10.1128/JB.180.19.5094-5101.1998
Filamentous bacteriophages of Vibrio parahaemolyticus as a possible clue to genetic transmission
Abstract
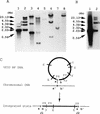
We have previously reported the isolation and characterization of two filamentous bacteriophages of Vibrio parahaemolyticus, designated Vf12 and Vf33. In this study, to understand the potential of these phages as tools for genetic transmission, we investigated the gene structures of replicative-form (RF) DNAs of their genomes and the distribution of these DNAs on chromosomal and extrachromosomal DNAs. The 7,965-bp nucleotide sequences of Vf12 and Vf33 were determined. An analysis of the overall gene structures revealed that Vf12 and Vf33 had conserved regions and distinctive regions. The gene organization of their conserved regions was similar to that of CTX phage of Vibrio cholerae and coliphage Ff of Escherichia coli, while their distinctive regions were characteristic of Vf12 and Vf33 phage genomes. Southern blot hybridization testing revealed that the filamentous phage genomes integrated into chromosomal DNA of V. parahaemolyticus at the distinctive region of the phage genome and were also distributed on some plasmids of V. parahaemolyticus and total cellular DNAs of one Vibrio damsela and one nonagglutinable Vibrio strain tested. These results strongly suggest the possibilities of genetic interaction among the bacteriophage Vf12 and Vf33 genomes and chromosomal and plasmid-borne DNAs of V. parahaemolyticus strains and of genetic transmission among strains through these filamentous phages.
Figures



References
-
- Baba K, Shirai H, Terai A, Kumagai K, Takeda Y, Nishibuchi M. Similarity of the tdh gene-bearing plasmids of Vibrio cholerae non-O1 and Vibrio parahaemolyticus. Microb Pathog. 1991;10:61–70. - PubMed
-
- Beck E, Zink B. Nucleotide sequence and genome organization of filamentous bacteriophages f1 and fd. Gene. 1981;16:35–58. - PubMed
-
- Blake P A, Weaver R E, Hollis D G. Diseases of humans (other than cholera) caused by vibrios. Annu Rev Microbiol. 1980;34:341–367. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

