Formation of single-stranded DNA during DNA transformation of Neisseria gonorrhoeae
- PMID: 9748444
- PMCID: PMC107547
- DOI: 10.1128/JB.180.19.5117-5122.1998
Formation of single-stranded DNA during DNA transformation of Neisseria gonorrhoeae
Abstract
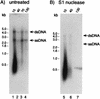
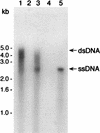
Neisseria gonorrhoeae is naturally competent for DNA transformation. In contrast to other natural prokaryotic DNA transformation systems, single-stranded donor DNA (ssDNA) has not previously been detected during transformation of N. gonorrhoeae. We have reassessed the physical nature of gonococcal transforming DNA by using a sensitive nondenaturing native blotting technique that detects ssDNA. Consistent with previous analyses, we found that the majority of donor DNA remained in the double-stranded form, and only plasmid DNAs that carried the genus-specific DNA uptake sequence were sequestered in a DNase I-resistant state. However, when the DNA was examined under native conditions, S1 nuclease-sensitive ssDNA was identified in all strains tested except for those bacteria that carried the dud-1 mutation. Surprisingly, ssDNA was also found during transformation of N. gonorrhoeae comA mutants, which suggested that ssDNA was initially formed within the periplasm.
Figures


References
-
- Danner D B, Deich R A, Sisco K L, Smith H O. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene. 1980;11:311–318. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

