Nucleotide sequence and characterization of cdrA, a cell division-related gene of Helicobacter pylori
- PMID: 9748467
- PMCID: PMC107570
- DOI: 10.1128/JB.180.19.5263-5268.1998
Nucleotide sequence and characterization of cdrA, a cell division-related gene of Helicobacter pylori
Abstract
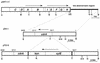
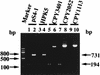
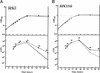
We identified cell division-related gene cdrA in Helicobacter pylori HPK5. The putative gene product, CdrA, is a 367-amino-acid polypeptide that exhibited a high level of homology to conserved hypothetical ATP-binding protein HP0066 of H. pylori 26695, except in the N-terminal region, and showed some similarity to the FtsK/SpoIIIE family proteins. We isolated a cdrA-disrupted mutant by allelic exchange mutagenesis. Because of the low transformation frequency, the possibility that a suppressing mutation would be found in the obtained cdrA mutant was discussed. A repressive role for CdrA on cell division was suggested by the observations that the wild-type strain formed filamentous cells in a high-salt level medium at early stationary phase, while a cdrA-disrupted mutant did not show such an abnormality. In addition, the wild-type strain adopted coccoid forms in the stationary phase, whereas the cdrA-disrupted mutant remained mostly as short rods. Furthermore, the cdrA-disrupted mutant regained the filamentation phenotype when the intact cdrA gene was introduced by allelic exchange. Taken together, these observations show that the cdrA gene plays an important role in the cell growth of H. pylori.
Figures


Similar articles
-
Cell elongation and cell death of helicobacter pylori is modulated by the disruption of cdrA (cell division-related gene A).Microbiol Immunol. 2006;50(7):487-97. doi: 10.1111/j.1348-0421.2006.tb03819.x. Microbiol Immunol. 2006. PMID: 16858140
-
Effect of Helicobacter pylori cdrA on interleukin-8 secretions and nuclear factor kappa B activation.World J Gastroenterol. 2012 Feb 7;18(5):425-34. doi: 10.3748/wjg.v18.i5.425. World J Gastroenterol. 2012. PMID: 22346248 Free PMC article.
-
Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange.J Bacteriol. 1993 Jun;175(11):3278-88. doi: 10.1128/jb.175.11.3278-3288.1993. J Bacteriol. 1993. PMID: 8501031 Free PMC article.
-
Cloning and characterization of the Helicobacter pylori flbA gene, which codes for a membrane protein involved in coordinated expression of flagellar genes.J Bacteriol. 1997 Feb;179(4):987-97. doi: 10.1128/jb.179.4.987-997.1997. J Bacteriol. 1997. PMID: 9023175 Free PMC article.
-
Cloning and functional characterization of the genes encoding 3-dehydroquinate synthase (aroB) and tRNA-guanine transglycosylase (tgt) from Helicobacter pylori.Med Microbiol Immunol. 1997 Oct;186(2-3):125-34. doi: 10.1007/s004300050054. Med Microbiol Immunol. 1997. PMID: 9403840
Cited by
-
Cloning, polymorphism, and inhibition of beta-carbonic anhydrase of Helicobacter pylori.J Gastroenterol. 2008;43(11):849-57. doi: 10.1007/s00535-008-2240-3. Epub 2008 Nov 18. J Gastroenterol. 2008. PMID: 19012038
-
Optimizing the growth of stressed Helicobacter pylori.J Microbiol Methods. 2011 Feb;84(2):174-82. doi: 10.1016/j.mimet.2010.11.015. Epub 2010 Dec 1. J Microbiol Methods. 2011. PMID: 21129415 Free PMC article.
-
Effect of host species on recG phenotypes in Helicobacter pylori and Escherichia coli.J Bacteriol. 2004 Nov;186(22):7704-13. doi: 10.1128/JB.186.22.7704-7713.2004. J Bacteriol. 2004. PMID: 15516585 Free PMC article.
-
Sodium chloride affects Helicobacter pylori growth and gene expression.J Bacteriol. 2008 Jun;190(11):4100-5. doi: 10.1128/JB.01728-07. Epub 2008 Mar 28. J Bacteriol. 2008. PMID: 18375562 Free PMC article.
-
Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape.PLoS Pathog. 2006 Sep;2(9):e97. doi: 10.1371/journal.ppat.0020097. PLoS Pathog. 2006. PMID: 17002496 Free PMC article.
References
-
- Blaser M J. Helicobacter pylori: microbiology of a “slow” bacterial infection. Trends Microbiol. 1993;1:255–260. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

