ATP hydrolysis catalyzed by human replication factor C requires participation of multiple subunits
- PMID: 9751713
- PMCID: PMC21688
- DOI: 10.1073/pnas.95.20.11607
ATP hydrolysis catalyzed by human replication factor C requires participation of multiple subunits
Abstract
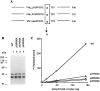
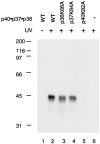
Human replication factor C (hRFC) is a five-subunit protein complex (p140, p40, p38, p37, and p36) that acts to catalytically load proliferating cell nuclear antigen onto DNA, where it recruits DNA polymerase delta or epsilon to the primer terminus at the expense of ATP, leading to processive DNA synthesis. We have previously shown that a subcomplex of hRFC consisting of three subunits (p40, p37, and p36) contained DNA-dependent ATPase activity. However, it is not clear which subunit(s) hydrolyzes ATP, as all five subunits include potential ATP binding sites. In this report, we introduced point mutations in the putative ATP-binding sequences of each hRFC subunit and examined the properties of the resulting mutant hRFC complex and the ATPase activity of the hRFC or the p40.p37.p36 complex. A mutation in any one of the ATP binding sites of the p36, p37, p40, or p140 subunits markedly reduced replication activity of the hRFC complex and the ATPase activity of the hRFC or the p40.p37.p36 complex. A mutation in the ATP binding site of the p38 subunit did not alter the replication activity of hRFC. These findings indicate that the replication activity of hRFC is dependent on efficient ATP hydrolysis contributed to by the action of four hRFC subunits.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

