The common marmoset: a new world primate species with limited Mhc class II variability
- PMID: 9751736
- PMCID: PMC21711
- DOI: 10.1073/pnas.95.20.11745
The common marmoset: a new world primate species with limited Mhc class II variability
Abstract
The common marmoset (Callithrix jacchus) is a New World primate species that is highly susceptible to fatal infections caused by various strains of bacteria. We present here a first step in the molecular characterization of the common marmoset's Mhc class II genes by nucleotide sequence analysis of the polymorphic exon 2 segments. For this study, genetic material was obtained from animals bred in captivity as well as in the wild. The results demonstrate that the common marmoset has, like other primates, apparently functional Mhc-DR and -DQ regions, but the Mhc-DP region has been inactivated. At the -DR and -DQ loci, only a limited number of lineages were detected. On the basis of the number of alleles found, the -DQA and -B loci appear to be oligomorphic, whereas only a moderate degree of polymorphism was observed for two of three Mhc-DRB loci. The contact residues in the peptide-binding site of the Caja-DRB1*03 lineage members are highly conserved, whereas the -DRB*W16 lineage members show more divergence in that respect. The latter locus encodes five oligomorphic lineages whose members are not observed in any other primate species studied, suggesting rapid evolution, as illustrated by frequent exchange of polymorphic motifs. All common marmosets tested were found to share one monomorphic type of Caja-DRB*W12 allele probably encoded by a separate locus. Common marmosets apparently lack haplotype polymorphism because the number of Caja-DRB loci present per haplotype appears to be constant. Despite this, however, an unexpectedly high number of allelic combinations are observed at the haplotypic level, suggesting that Caja-DRB alleles are exchanged frequently between chromosomes by recombination, promoting an optimal distribution of limited Mhc polymorphisms among individuals of a given population. This peculiar genetic make up, in combination with the limited variability of the major histocompatability complex class II repertoire, may contribute to the common marmoset's susceptibility to particular bacterial infections.
Figures

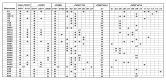

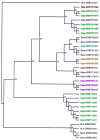
References
-
- Kaufman J, Skjoedt K, Salomonsen J. Immunol Rev. 1990;113:83–117. - PubMed
-
- Parham P, Ohta T. Science. 1996;272:67–74. - PubMed
-
- Klein J, Satta Y, O’hUigin C, Takahata N. Annu Rev Immunol. 1993;11:269–295. - PubMed
-
- Bontrop R E, Otting N, Slierendregt B L, Lanchbury J S. Immunol Rev. 1995;143:33–62. - PubMed
-
- Klein J, O’hUigin C. Immunol Rev. 1995;143:89–111. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

