A bioluminescence assay using Nitrosomonas europaea for rapid and sensitive detection of nitrification inhibitors
- PMID: 9758781
- PMCID: PMC106494
- DOI: 10.1128/AEM.64.10.3656-3662.1998
A bioluminescence assay using Nitrosomonas europaea for rapid and sensitive detection of nitrification inhibitors
Abstract
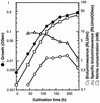
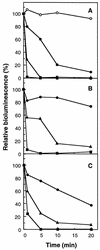
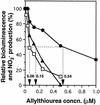
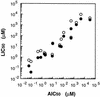
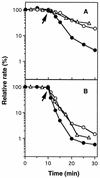
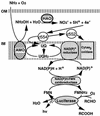
An expression vector for the luxAB genes, derived from Vibrio harveyi, was introduced into Nitrosomonas europaea. Although the recombinant strain produced bioluminescence due to the expression of the luxAB genes under normal growing conditions, the intensity of the light emission decreased immediately, in a time-and dose-dependent manner, with the addition of ammonia monooxygenase inhibitors, such as allylthiourea, phenol, and nitrapyrin. When whole cells were challenged with several nitrification inhibitors and toxic compounds, a close relationship was found between the change in the intensity of the light emission and the level of ammonia-oxidizing activity. The response of bioluminescence to the addition of allylthiourea was considerably faster than the change in the ammonia-oxidizing rate, measured as both the O2 uptake and NO2- production rates. The bioluminescence of cells inactivated by ammonia monooxygenase inhibitor was recovered rapidly by the addition of certain substrates for hydroxylamine oxidoreductase. These results suggested that the inhibition of bioluminescence was caused by the immediate decrease of reducing power in the cell due to the inactivation of ammonia monooxygenase, as well as by the destruction of other cellular metabolic pathways. We conclude that the assay system using luminous Nitrosomonas can be applied as a rapid and sensitive detection test for nitrification inhibitors, and it will be used to monitor the nitrification process in wastewater treatment plants.
Figures
References
-
- Aleem M I H. Generation of reducing power in chemosynthesis. II. Energy linked reduction of pyridine nucleotide in the chemoautotroph Nitrosomonas europaea. Biochim Biophys Acta. 1966;113:216–224. - PubMed
-
- Berks B C, Ferguson S J, Moir J W B, Richardson D J. Enzyme and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. - PubMed
-
- Bulich A A, Greene M W, Isenberg D L. A practical and reliable method for monitoring the toxicity of aquatic samples. Process Biochem. 1982;17:45–47.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

