Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples
- PMID: 9758798
- PMCID: PMC106545
- DOI: 10.1128/AEM.64.10.3769-3775.1998
Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples
Abstract
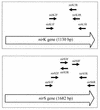
A system was developed for the detection of denitrifying bacteria by the amplification of specific nitrite reductase gene fragments with PCR. Primer sequences were found for the amplification of fragments from both nitrite reductase genes (nirK and nirS) after comparative sequence analysis. Whenever amplification was tried with these primers, the known nir type of denitrifying laboratory cultures could be confirmed. Likewise, the method allowed a determination of the nir type of five laboratory strains. The nirK gene could be amplified from Blastobacter denitrificans, Alcaligenes xylosoxidans, and Alcaligenes sp. (DSM 30128); the nirS gene was amplified from Alcaligenes eutrophus DSM 530 and from the denitrifying isolate IFAM 3698. For each of the two genes, at least one primer combination amplified successfully for all of the test strains. Specific amplification products were not obtained with nondenitrifying bacteria or with strains of the other nir type. The specificity of the amplified products was confirmed by subsequent sequencing. These results suggest the suitability of the method for the qualitative detection of denitrifying bacteria in environmental samples. This was shown by applying one generally amplifying primer combination for each nir gene developed in this study to total DNA preparations from aquatic habitats.
Figures
References
-
- Ausubel F M, Brent R, Kinston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991.
-
- Betlach M R. Evolution of bacterial denitrification and denitrifier diversity. Antonie Leeuwenhoek. 1982;48:585–607. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

