Enzymatic conversion of glucose to UDP-4-keto-6-deoxyglucose in Streptomyces spp
- PMID: 9758828
- PMCID: PMC106587
- DOI: 10.1128/AEM.64.10.3972-3976.1998
Enzymatic conversion of glucose to UDP-4-keto-6-deoxyglucose in Streptomyces spp
Abstract
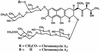
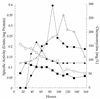
All of the 2,6-dideoxy sugars contained within the structure of chromomycin A3 are derived from D-glucose. Enzyme assays were used to confirm the presence of hexokinase, phosphoglucomutase, UDPG pyrophosphorylase (UDPGP), and UDPG oxidoreductase (UDPGO), all of which are involved in the pathway of glucose activation and conversion into 2,6-dideoxyhexoses during chromomycin biosynthesis. Levels of the four enzymes in Streptomyces spp. cell extracts were correlated with the production of chromomycins. The pathway of sugar activation in Streptomyces spp. involves glucose 6-phosphorylation by hexokinase, isomerization to G-1-P catalyzed by phosphoglucomutase, synthesis of UDPG catalyzed by UDPGP, and formation of UDP-4-keto-6-deoxyglucose by UDPGO.
Figures

References
-
- Becker A, Kleickmann A, Keller M, Arnold W, Puhler A. Identification and analysis of the Rhizobium meliloti exo AMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exo HKLAMONP fragment. Mol Gen Genet. 1993;241:367–379. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Chang H, Lee C, Peng H. Identification of the Pseudomonas UDPG pyrophosphorylase gene. 1993. From NCBI (accession no. U03751).
-
- Elbein A D, Heath E C. The biosynthesis of cell wall lipopolysaccharide in Escherichia coli. II. Guanosine diphosphate 4-keto-6-deoxy-d-mannose, an intermediate in the biosynthesis of guanosine diphosphate colitose. J Biol Chem. 1965;240:1926–1931. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

