Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1
- PMID: 9763420
- PMCID: PMC2132809
- DOI: 10.1083/jcb.143.1.49
Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1
Abstract
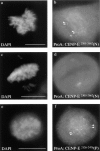
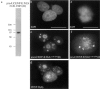
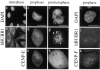
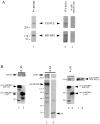
We have identified a 350-amino acid domain in the kinetochore motor CENP-E that specifies kinetochore binding in mitosis but not during interphase. The kinetochore binding domain was used in a yeast two-hybrid screen to isolate interacting proteins that included the kinetochore proteins CENP-E, CENP-F, and hBUBR1, a BUB1-related kinase that was found to be mutated in some colorectal carcinomas (Cahill, D.P., C. Lengauer, J. Yu, G.J. Riggins, J.K. Wilson, S.D. Markowitz, K.W. Kinzler, and B. Vogelstein. 1998. Nature. 392:300-303). CENP-F, hBUBR1, and CENP-E assembled onto kinetochores in sequential order during late stages of the cell cycle. These proteins therefore define discrete steps along the kinetochore assembly pathway. Kinetochores of unaligned chromosome exhibited stronger hBUBR1 and CENP-E staining than those of aligned chromosomes. CENP-E and hBUBR1 remain colocalized at kinetochores until mid-anaphase when hBUBR1 localized to portions of the spindle midzone that did not overlap with CENP-E. As CENP-E and hBUBR1 can coimmunoprecipitate with each other from HeLa cells, they may function as a motor-kinase complex at kinetochores. However, the complex distribution pattern of hBUBR1 suggests that it may regulate multiple functions that include the kinetochore and the spindle midzone.
Figures







References
-
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Wilson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. - PubMed
-
- Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

