Coilin can form a complex with the U7 small nuclear ribonucleoprotein
- PMID: 9763457
- PMCID: PMC25576
- DOI: 10.1091/mbc.9.10.2987
Coilin can form a complex with the U7 small nuclear ribonucleoprotein
Abstract
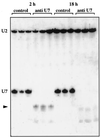
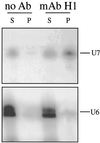
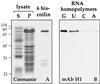
Coiled bodies (CBs) in the amphibian oocyte nucleus are spherical structures up to 10 microm or more in diameter, much larger than their somatic counterparts, which rarely exceed 1 microm. Oocyte CBs may have smaller granules attached to their surface or embedded within them, which are identical in structure and composition to the many hundreds of B-snurposomes found free in the nucleoplasm. The matrix of the CBs contains the diagnostic protein p80-coilin, which is colocalized with the U7 small nuclear ribonucleoprotein (snRNP), whereas the attached and embedded B-snurposomes contain splicing snRNPs. A few of the 50-100 CBs in the oocyte nucleus are attached to lampbrush chromosomes at the histone gene loci. By coimmunoprecipitation we show that coilin and the U7 snRNP can form a weak but specific complex in the nucleoplasm, which is dependent on the special U7 Sm-binding site. Under the same conditions coilin does not associate with the U1 and U2 snRNPs. Coilin is a nucleic acid-binding protein, as shown by its interaction with single-stranded DNA and with poly r(U) and poly r(G). We suggest that an important function of coilin is to form a transient complex with the U7 snRNP and accompany it to the CBs. In the case of CBs attached to chromosomes at the histone gene loci, the U7 snRNP is thus brought close to the actual site of histone pre-mRNA transcription.
Figures






References
-
- Bandziulis RJ, Swanson MS, Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

