Regulation of L-selectin-mediated rolling through receptor dimerization
- PMID: 9763619
- PMCID: PMC2212497
- DOI: 10.1084/jem.188.7.1385
Regulation of L-selectin-mediated rolling through receptor dimerization
Abstract
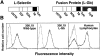
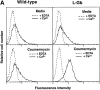
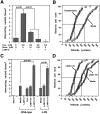
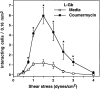
L-selectin binding activity for its ligand expressed by vascular endothelium is rapidly and transiently increased after leukocyte activation. To identify mechanisms for upregulation and assess how this influences leukocyte/endothelial cell interactions, cell-surface dimers of L-selectin were induced using the coumermycin-GyrB dimerization strategy for cross-linking L-selectin cytoplasmic domains in L-selectin cDNA-transfected lymphoblastoid cells. Coumermycin- induced L-selectin dimerization resulted in an approximately fourfold increase in binding of phosphomanan monoester core complex (PPME), a natural mimic of an L-selectin ligand, comparable to that observed after leukocyte activation. Moreover, L-selectin dimerization significantly increased (by approximately 700%) the number of lymphocytes rolling on vascular endothelium under a broad range of physiological shear stresses, and significantly slowed their rolling velocities. Therefore, L-selectin dimerization may explain the rapid increase in ligand binding activity that occurs after leukocyte activation and may directly influence leukocyte migration to peripheral lymphoid tissues or to sites of inflammation. Inducible oligomerization may also be a common mechanism for rapidly upregulating the adhesive or ligand-binding function of other cell-surface receptors.
Figures

References
-
- Rosen SD, Bertozzi CR. The selectins and their ligands. Curr Opin Cell Biol. 1994;6:663–673. - PubMed
-
- Tedder TF, Steeber DA, Chen A, Engel P. The selectins: vascular adhesion molecules. FASEB (Fed Am Soc Exp Biol) J. 1995;9:866–873. - PubMed
-
- Spertini O, Kansas GS, Munro JM, Griffin JD, Tedder TF. Regulation of leukocyte migration by activation of the leukocyte adhesion molecule-1 (LAM-1) selectin. Nature. 1991;349:691–694. - PubMed
-
- Haribabu B, Steeber DA, Ali H, Richardson RM, Snyderman R, Tedder TF. Chemoattractant receptor-induced phosphorylation of L-selectin. J Biol Chem. 1997;272:13961–13965. - PubMed
-
- Picker LJ, Warnock RA, Burns AR, Doerschuk CM, Berg EL, Butcher EC. The neutrophil selectin LECAM-1 presents carbohydrate ligands to the vascular selectins ELAM-1 and GMP-140. Cell. 1991;66:921–933. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

