Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein
- PMID: 9765205
- PMCID: PMC317192
- DOI: 10.1101/gad.12.19.3032
Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein
Abstract
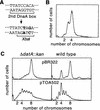
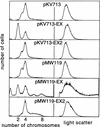
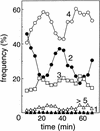
Replication of the Escherichia coli chromosome is initiated at a unique site, oriC. Concurrent initiation occurs at all oriC sites present in a cell once, and only once, per cell cycle. A mechanism to ensure cyclic initiation events was found operating through the chromosomal site, datA, a 1-kb segment located at 94.7 min on the genetic map that titrates exceptionally large amounts of the bacterial initiator protein, DnaA. A strain lacking datA grew normally but exhibited an asynchronous initiation phenotype as a result of extra initiation events. This mutant phenotype was suppressed by DnaA-titrating plasmids. Furthermore, mutations in a 9-bp DnaA-binding sequence (the DnaA box) in datA were enough to induce the mutant phenotype. Thus, datA is a novel chromosomal element that appears to adjust a balance between free and bound DnaA for a single initiation event at a fixed time in the bacterial cell cycle. Titration of DnaA to newly duplicated datA during oriC sequestration, which is mediated by hemimethylated GATC sequences in oriC and the SeqA protein, would contribute to prevention of reinitiations when oriC is desequestered.
Figures




References
-
- Bird RE, Louarn J, Martuscelli J, Caro L. Origin and sequence of chromosome replication in Escherichia coli. J Mol Biol. 1972;70:549–566. - PubMed
-
- Boye E, Løbner-Olesen A. The role of dam methyltransferase in the control of DNA replication in E. coli. Cell. 1990;62:981–989. - PubMed
-
- Boye E, Løbner-Olesen A, Skarstad K. Timing of chromosomal replication in Escherichia coli. Biochim Biophys Acta. 1988;951:359–364. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
