Synergistic cooperation of TFE3 and smad proteins in TGF-beta-induced transcription of the plasminogen activator inhibitor-1 gene
- PMID: 9765209
- PMCID: PMC317197
- DOI: 10.1101/gad.12.19.3084
Synergistic cooperation of TFE3 and smad proteins in TGF-beta-induced transcription of the plasminogen activator inhibitor-1 gene
Abstract
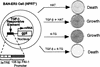
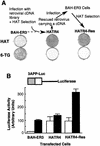
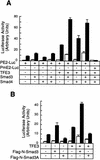
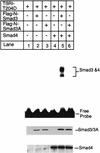
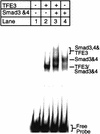
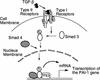
Members of the TGF-beta superfamily influence a broad range of biological activities including stimulation of wound healing and inhibition of cell growth. TGF-beta signals through type I and II receptor serine/ threonine kinases and induces transcription of many genes including plasminogen activator inhibitor-1 (PAI-1). To identify proteins that participate in TGF-beta-induced gene expression, we developed a novel retrovirus-mediated expression cloning strategy; and using this approach, we established that transcription factor microE3 (TFE3) is involved in TGF-beta-induced activation of the PAI-1 promoter. We showed that TFE3 binds to an E-box sequence in PE2, a 56-bp promoter fragment of the PAI-1 promoter, and that mutation of this sequence abolishes both TFE3 binding as well as TGF-beta-dependent activation. TFE3 and Smad3 synergistically activate the PE2 promoter and phosphorylated Smad3 and Smad4 bind to a sequence adjacent to the TFE3-binding site in this promoter. Binding of both TFE3 and the Smad proteins to their cognate sequences is indispensable for TGF-beta-inducible activation of the PE2 promoter. Hence, TFE3 is an important transcription factor in at least one TGF-beta-activated signal transduction pathway.
Figures
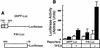





References
-
- Abdollah S, Macias-Silva M, Tzukazaki T, Hayashi H, Attisano L, Wrana J. TβRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2–Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. - PubMed
-
- Attisano L, Wrana J. Mads and Smads in TGFβ signaling. Curr Opin Cell Biol. 1998;10:188–194. - PubMed
-
- Attisano L, Wrana JL, Lopez-Casillas F, Massagué J. TGF-β receptors and actions. Biochimica et Biophysica Acta. 1994;1222:71–80. - PubMed
-
- Beckmann H, Su L-K, Kadesch T. TFE3: A helix-loop-helix protein that activates transcription through the immunoglobulin enhancer μE3 motif. Genes & Dev. 1990;4:167–179. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
