Interaction of a nascent RNA structure with RNA polymerase is required for hairpin-dependent transcriptional pausing but not for transcript release
- PMID: 9765211
- PMCID: PMC317188
- DOI: 10.1101/gad.12.19.3110
Interaction of a nascent RNA structure with RNA polymerase is required for hairpin-dependent transcriptional pausing but not for transcript release
Abstract
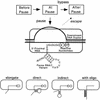
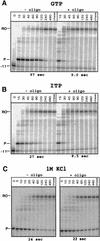
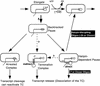
Nascent RNA structures may regulate RNA chain elongation either directly through interaction with RNA polymerase or indirectly by disrupting nascent RNA contacts with polymerase or DNA. To distinguish these mechanisms we tested whether the effects of the his leader pause RNA hairpin could be mimicked by pairing of antisense DNA or RNA oligonucleotides to the nascent transcript. The his pause hairpin inhibits nucleotide addition when it forms 11 nucleotides from the transcript 3' end. It also can terminate transcription when base changes extend its stem to </=8 nucleotides from the 3' end. All oligonucleotides that disrupted the pause hairpin reduced the dwell time of RNA polymerase at the pause site dramatically, even when they mimicked the 11-nucleotide 3'-proximal RNA spacing or created a suitably positioned RNA loop. Oligonucleotides that paired </=8 nucleotides from the pause RNA 3' end could trigger transcript release, but only when added to an already paused complex. These results argue that direct interaction of a nascent RNA hairpin with RNA polymerase delays escape from a pause, but that indirect effects of a hairpin may trigger transcript release from a paused complex. Resistance of the paused complex to pyrophosphorolysis and its reversal by antisense oligonucleotides further suggest that interaction of the pause hairpin with RNA polymerase disengages the RNA 3' end from the active site.
Figures






References
-
- Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. - PubMed
-
- Burgess RR, Jendrisak JJ. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975;14:4634–4638. - PubMed
-
- Chamberlin MJ. The Harvey lectures. New York, NY: Wiley-Liss; 1995. New models for the mechanism of transcription elongation and its regulation; pp. 1–21. - PubMed
-
- Chan C, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993;233:25–42. - PubMed
-
- ————— Effects of neutral salts of transcript elongation and pausing suggest the his leader pause RNA hairpin interacts with an easily disordered region of RNA polymerase. J Mol Biol. 1997;268:37–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
