Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent In situ hybridization
- PMID: 9765396
- PMCID: PMC110268
- DOI: 10.1128/JVI.72.11.8578-8585.1998
Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent In situ hybridization
Abstract
The time courses of poliovirus plus- and minus-strand RNA synthesis in infected HEp-2 cells were monitored separately, using a quantitative RNase assay. In parallel, viral RNA and proteins were located in situ by confocal microscopy within cells fixed by a protocol determined to retain their native size and shape. Plus- and minus-strand RNAs were visualized by fluorescent in situ hybridization (FISH) with strand-specific riboprobes. The probes were labelled with different fluorochromes to allow for the simultaneous detection of plus- and minus-strand RNA. The FISH experiments showed minus-strand RNA to be present in distinct, regularly sized, round structures throughout the viral replication cycle. Plus-strand RNA was found in the same structures and also in smaller clusters of vesicles. Association of viral RNA with membranes was demonstrated by combining FISH with immunofluorescence (IF) detection of the viral 2B- and 2C-containing P2 proteins, which are known to be markers for virus-induced membranes. At early times postinfection, the virus-induced membranous structures were distributed through most of the cytoplasm, whereas around peak RNA synthesis, both RNA-associated membranous structures migrated to the center of the cell. During this process, the plus- and minus-strand-containing larger structures stayed as recognizable entities, whereas the plus-strand-containing granules coalesced into a juxtanuclear area of membranous vesicles. An involvement of Golgi-derived membranes in the formation of virus-induced vesicles and RNA synthesis early in infection was investigated by IF with 2C- and Golgi-specific antibodies.
Figures


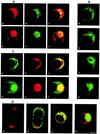

References
-
- Aldabe R, Barco A, Carrasco L. Membrane permeabilization by poliovirus proteins 2B and 2BC. J Biol Chem. 1996;271:23134–23137. - PubMed
-
- Aldabe R, Carrasco L. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem Biophys Res Commun. 1995;206:64–76. - PubMed
-
- Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. - PubMed
-
- Baron M H, Baltimore D. In vitro copying of viral positive strand RNA by poliovirus replicase. Characterization of the reaction and its products. J Biol Chem. 1982;257:12359–12366. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

