Ribosomal S27a coding sequences upstream of ubiquitin coding sequences in the genome of a pestivirus
- PMID: 9765411
- PMCID: PMC110283
- DOI: 10.1128/JVI.72.11.8697-8704.1998
Ribosomal S27a coding sequences upstream of ubiquitin coding sequences in the genome of a pestivirus
Abstract
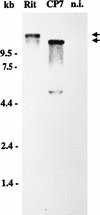
Molecular characterization of cytopathogenic (cp) bovine viral diarrhea virus (BVDV) strain CP Rit, a temperature-sensitive strain widely used for vaccination, revealed that the viral genomic RNA is about 15.2 kb long, which is about 2.9 kb longer than the one of noncytopathogenic (noncp) BVDV strains. Molecular cloning and nucleotide sequencing of parts of the genome resulted in the identification of a duplication of the genomic region encoding nonstructural proteins NS3, NS4A, and part of NS4B. In addition, a nonviral sequence was found directly upstream of the second copy of the NS3 gene. The 3' part of this inserted sequence encodes an N-terminally truncated ubiquitin monomer. This is remarkable since all described cp BVDV strains with ubiquitin coding sequences contain at least one complete ubiquitin monomer. The 5' region of the nonviral sequence did not show any homology to cellular sequences identified thus far in cp BVDV strains. Databank searches revealed that this second cellular insertion encodes part of ribosomal protein S27a. Further analyses included molecular cloning and nucleotide sequencing of the cellular recombination partner. Sequence comparisons strongly suggest that the S27a and the ubiquitin coding sequences found in the genome of CP Rit were both derived from a bovine mRNA encoding a hybrid protein with the structure NH2-ubiquitin-S27a-COOH. Polyprotein processing in the genomic region encoding the N-terminal part of NS4B, the two cellular insertions, and NS3 was studied by a transient-expression assay. The respective analyses showed that the S27a-derived polypeptide, together with the truncated ubiquitin, served as processing signal to yield NS3, whereas the truncated ubiquitin alone was not capable of mediating the cleavage. Since the expression of NS3 is strictly correlated with the cp phenotype of BVDV, the altered genome organization leading to expression of NS3 most probably represents the genetic basis of cytopathogenicity of CP Rit.
Figures



Similar articles
-
RNA recombination between persisting pestivirus and a vaccine strain: generation of cytopathogenic virus and induction of lethal disease.J Virol. 2001 Jul;75(14):6256-64. doi: 10.1128/JVI.75.14.6256-6264.2001. J Virol. 2001. PMID: 11413291 Free PMC article.
-
Insertion of cellular NEDD8 coding sequences in a pestivirus.Virology. 2000 Dec 20;278(2):456-66. doi: 10.1006/viro.2000.0644. Virology. 2000. PMID: 11118368
-
Cytopathogenicity of a pestivirus correlates with a 27-nucleotide insertion.J Virol. 1996 Nov;70(11):7851-8. doi: 10.1128/JVI.70.11.7851-7858.1996. J Virol. 1996. PMID: 8892907 Free PMC article.
-
Insertion of cellular sequences in the genome of bovine viral diarrhea virus.Arch Virol Suppl. 1991;3:133-42. doi: 10.1007/978-3-7091-9153-8_15. Arch Virol Suppl. 1991. PMID: 9210934 Review.
-
Pathogenesis of mucosal disease, a deadly disease of cattle caused by a pestivirus.Clin Diagn Virol. 1998 Jul 15;10(2-3):121-7. doi: 10.1016/s0928-0197(98)00037-3. Clin Diagn Virol. 1998. PMID: 9741637 Review.
Cited by
-
Cytopathogenicity of classical Swine Fever virus correlates with attenuation in the natural host.J Virol. 2008 Oct;82(19):9717-29. doi: 10.1128/JVI.00782-08. Epub 2008 Jul 23. J Virol. 2008. PMID: 18653456 Free PMC article.
-
Nonhomologous RNA recombination in bovine viral diarrhea virus: molecular characterization of a variety of subgenomic RNAs isolated during an outbreak of fatal mucosal disease.J Virol. 1999 Jul;73(7):5646-53. doi: 10.1128/JVI.73.7.5646-5653.1999. J Virol. 1999. PMID: 10364314 Free PMC article.
-
Elimination of Foreign Sequences in Eukaryotic Viral Reference Genomes Improves the Accuracy of Virome Analysis.mSystems. 2022 Dec 20;7(6):e0090722. doi: 10.1128/msystems.00907-22. Epub 2022 Oct 26. mSystems. 2022. PMID: 36286492 Free PMC article.
-
Noncytopathogenic pestivirus strains generated by nonhomologous RNA recombination: alterations in the NS4A/NS4B coding region.J Virol. 2005 Nov;79(22):14261-70. doi: 10.1128/JVI.79.22.14261-14270.2005. J Virol. 2005. PMID: 16254361 Free PMC article.
-
Viral proteins acquired from a host converge to simplified domain architectures.PLoS Comput Biol. 2012 Feb;8(2):e1002364. doi: 10.1371/journal.pcbi.1002364. Epub 2012 Feb 2. PLoS Comput Biol. 2012. PMID: 22319434 Free PMC article.
References
-
- Agol V I. Recombination and other genomic rearrangements in picornaviruses. Semin Virol. 1997;8:77–84.
-
- Baker J C. Bovine viral diarrhea virus: a review. J Am Vet Med Assoc. 1987;190:1449–1458. - PubMed
-
- Becher, P. Unpublished data.
-
- Becher P, König M, Paton D, Thiel H-J. Further characterization of border disease virus isolates: evidence for the presence of more than three species within the genus pestivirus. Virology. 1995;209:200–206. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

