Isolation of an Arabidopsis thaliana mutant in which the multiplication of both cucumber mosaic virus and turnip crinkle virus is affected
- PMID: 9765416
- PMCID: PMC110288
- DOI: 10.1128/JVI.72.11.8731-8737.1998
Isolation of an Arabidopsis thaliana mutant in which the multiplication of both cucumber mosaic virus and turnip crinkle virus is affected
Erratum in
- J Virol. 2003 Jul;77(14):8178
Abstract
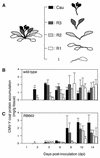
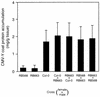
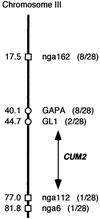
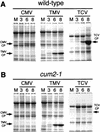
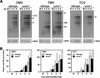
During the systemic infection of plants by viruses, host factors play an important role in supporting virus multiplication. To identify and characterize the host factors involved in this process, we isolated an Arabidopsis thaliana mutant named RB663, in which accumulation of the coat protein (CP) of cucumber mosaic virus (CMV) in upper uninoculated leaves was delayed. Genetic analyses suggested that the phenotype of delayed accumulation of CMV CP in RB663 plants was controlled by a monogenic, recessive mutation designated cum2-1, which is located on chromosome III and is distinct from the previously characterized cum1 mutation. Multiplication of CMV was delayed in inoculated leaves of RB663 plants, whereas the multiplication in RB663 protoplasts was similar to that in wild-type protoplasts. This suggests that the cum2-1 mutation affects the cell-to-cell movement of CMV rather than CMV replication within a single cell. In RB663 plants, the multiplication of turnip crinkle virus (TCV) was also delayed but that of tobacco mosaic virus was not affected. As observed with CMV, the multiplication of TCV was normal in protoplasts and delayed in inoculated leaves of RB663 plants compared to that in wild-type plants. Furthermore, the phenotype of delayed TCV multiplication cosegregated with the cum2-1 mutation as far as we examined. Therefore, the cum2-1 mutation is likely to affect the cell-to-cell movement of both CMV and TCV, implying a common aspect to the mechanisms of cell-to-cell movement in these two distinct viruses.
Figures
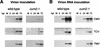
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994.
-
- Bell C J, Ecker J R. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. - PubMed
-
- Boccard F, Baulcombe D. Mutational analysis of cis-acting sequences and gene function in RNA3 of cucumber mosaic virus. Virology. 1993;193:563–578. - PubMed
-
- Camilleri C, Lafleuriel J, Macadre C, Varoquaux F, Parmentier Y, Picard G, Caboche M, Bouchez D. A YAC contig map of Arabidopsis thaliana chromosome 3. Plant J. 1998;14:633–642. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

