CD4 promoter transactivation by human herpesvirus 6
- PMID: 9765424
- PMCID: PMC110296
- DOI: 10.1128/JVI.72.11.8797-8805.1998
CD4 promoter transactivation by human herpesvirus 6
Abstract
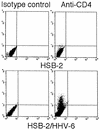
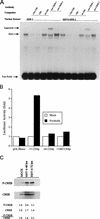
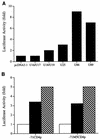
The observation that human herpesvirus 6 (HHV-6) can induce CD4 gene transcription and expression in CD4(-) cells was reported several years ago (P. Lusso, A. De Maria, M. Malnati, F. Lori, S. E. DeRocco, M. Baseler, and R. C. Gallo, Nature 349:533-535, 1991) and subsequently confirmed (P. Lusso, M. S. Malnati, A. Garzino-Demo, R. W. Crowley, E. O. Long, and R. C. Gallo, Nature 362:458-462, 1993; G. Furlini, M. Vignoli, E. Ramazzotti, M. C. Re, G. Visani, and M. LaPlaca, Blood 87:4737-4745, 1996). Our objective was to identify the mechanisms underlying such phenomena. Using reporter gene constructs driven by the CD4 promoter, we report that HHV-6 can efficiently transactivate such genetic elements. Activation of the CD4 promoter occurs in the presence of the viral DNA polymerase inhibitor phosphonoformic acid, which limits expression to the immediate-early and early classes of viral genes. Using deletion mutants and specific CD4 promoter mutants, we identified an ATF/CRE binding site located at nucleotides -67 to -60 upstream of the CD4 gene transcription start site that is important for HHV-6 transactivation. The ATF/CRE site is also essential for CD4 promoter activation by forskolin, an activator of adenylate cyclase. Using electrophoretic mobility shift assays and specific antibodies, we showed that CREB-1 binds specifically to the -79 to -52 region of the CD4 promoter. Last, we have identified two open reading frames (ORFs) of HHV-6, U86 and U89 from the immediate-early locus A, that can transactivate the CD4 promoter in HeLa cells. However, transactivation of the CD4 promoter by ORFs U86 and U89 is independent of the CRE element, suggesting that additional HHV-6 ORFs are likely to contribute to CD4 gene activation. Taken together, our results will help to understand the complex interactions occurring between HHV-6 and the CD4 promoter and provide additional information regarding the class of transcription factors involved in the control of CD4 gene expression.
Figures


References
-
- Becker W B, Engelbrecht S, Becker M L, Piek C, Robson B A, Wood L, Jacobs P. Isolation of a new human herpesvirus producing a lytic infection of helper (CD4) T-lymphocytes in peripheral blood lymphocyte cultures—another cause of acquired immunodeficiency? S Afr Med J. 1988;74:610–614. - PubMed
-
- Blue M L, Daley J F, Levine H, Craig K A, Schlossman S F. Biosynthesis and surface expression of T8 by peripheral blood T4+ cells in vitro. J Immunol. 1986;137:1202–1207. - PubMed
-
- Cammarota G, Scheirle A, Takacs B, Doran D M, Knorr R, Bannwarth W, Guardiola J, Sinigaglia F. Identification of a CD4 binding site on the beta 2 domain of HLA-DR molecules. Nature. 1992;356:799–801. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

