In vivo footprinting of the enhancer sequences in the upstream long terminal repeat of Moloney murine leukemia virus: differential binding of nuclear factors in different cell types
- PMID: 9765441
- PMCID: PMC110313
- DOI: 10.1128/JVI.72.11.8961-8970.1998
In vivo footprinting of the enhancer sequences in the upstream long terminal repeat of Moloney murine leukemia virus: differential binding of nuclear factors in different cell types
Abstract
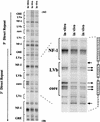
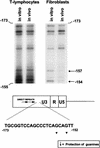
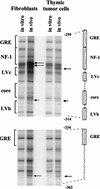
The enhancer sequences in the Moloney murine leukemia virus (M-MuLV) long terminal repeat (LTR) are of considerable interest since they are crucial for virus replication and the ability of the virus to induce T lymphomas. While extensive studies have identified numerous nuclear factors that can potentially bind to M-MuLV enhancer DNA in vitro, it has not been made clear which of these factors are bound in vivo. To address this problem, we carried out in vivo footprinting of the M-MuLV enhancer in infected cells by in vivo treatment with dimethyl sulfate (DMS) followed by visualization through ligation-mediated PCR (LMPCR) and gel electrophoresis. In vivo DMS-LMPCR footprinting of the upstream LTR revealed evidence for factor binding at several previously characterized motifs. In particular, protection of guanines in the central LVb/Ets and Core sites within the 75-bp repeats was detected in infected NIH 3T3 fibroblasts, Ti-6 lymphoid cells, and thymic tumor cells. In contrast, factor binding at the NF-1 sites was found in infected fibroblasts but not in T-lymphoid cells. These results are consistent with the results of previous experiments indicating the importance of the LVb/Ets and Core sequences for many retroviruses and the biological importance especially of the NF-1 sites in fibroblasts and T-lymphoid cells. No evidence for factor binding to the glucocorticoid responsive element and LVa sites was found. Additional sites of protein binding included a region in the GC-rich sequences downstream of the 75-bp repeats (only in fibroblasts), a hypersensitive guanine on the minus strand in the LVc site (only in T-lymphoid cells), and a region upstream of the 75-bp repeats. These experiments provide concrete evidence for the differential in vivo binding of nuclear factors to the M-MuLV enhancers in different cell types.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

