Molecular evidence for distinct genotypes of monkey B virus (herpesvirus simiae) which are related to the macaque host species
- PMID: 9765470
- PMCID: PMC110342
- DOI: 10.1128/JVI.72.11.9224-9232.1998
Molecular evidence for distinct genotypes of monkey B virus (herpesvirus simiae) which are related to the macaque host species
Abstract
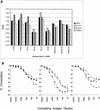
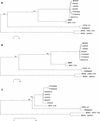
Although monkey B virus (herpesvirus simiae; BV) is common in all macaque species, fatal human infections appear to be associated with exposure to rhesus macaques (Macaca mulatta), suggesting that BV isolates from rhesus monkeys may be more lethal to nonmacaques than are BV strains indigenous to other macaque species. To determine if significant differences that would support this supposition exist among BV isolates, we compared multiple BV strains isolated from rhesus, cynomolgus, pigtail, and Japanese macaques. Antigenic analyses indicated that while the isolates were very closely related to one another, there are some antigenic determinants that are specific to BV isolates from different macaque species. Restriction enzyme digest patterns of viral DNA revealed marked similarities between rhesus and Japanese macaque isolates, while pigtail and cynomolgus macaque isolates had distinctive cleavage patterns. To further compare genetic diversity among BV isolates, DNA sequences from two regions of the viral genome containing genes that are conserved (UL27 and US6) and variable (US4 and US5) among primate alphaherpesviruses, as well as from two noncoding intergenic regions, were determined. From these sequence data and a phylogenetic analysis of them it was evident that while all isolates were closely related strains of BV, there were three distinct genotypes. The three BV genotypes were directly related to the macaque species of origin and were composed of (i) isolates from rhesus and Japanese macaques, (ii) cynomolgus monkey isolates, and (iii) isolates from pigtail macaques. This study demonstrates the existence of different BV genotypes which are related to the macaque host species and thus provides a molecular basis for the possible existence of BV isolates which vary in their levels of pathogenicity for nonmacaque species.
Figures




References
-
- Bennett A M, Harrington L, Kelly D C. Nucleotide sequence analysis of genes encoding glycoproteins D and J in simian herpes B virus. J Gen Virol. 1992;73:2963–2967. - PubMed
-
- Benson P M, Malane S L, Banks R, Hicks C B, Hilliard J K. B virus (Herpesvirus simiae) and human infection. Arch Dermatol. 1989;125:1247–1248. - PubMed
-
- Black D, Eberle R. Detection and differentiation of primate α-herpes-viruses by PCR. J Vet Diagn Investig. 1997;9:225–231. - PubMed
-
- Centers for Disease Control. B-virus infection in humans—Pensacola, Florida. Morbid Mortal Weekly Rep. 1987;36:289–290. , 295–296. - PubMed
-
- Davenport D S, Johnson D R, Holmes G P, Jewett D A, Ross S C, Hilliard J K. Diagnosis and management of human B virus (Herpesvirus simiae) infections in Michigan. Clin Infect Dis. 1994;19:33–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

