The 20S proteasome of Streptomyces coelicolor
- PMID: 9765579
- PMCID: PMC107596
- DOI: 10.1128/JB.180.20.5448-5453.1998
The 20S proteasome of Streptomyces coelicolor
Abstract
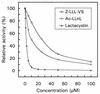
20S proteasomes were purified from Streptomyces coelicolor A3(2) and shown to be built from one alpha-type subunit (PrcA) and one beta-type subunit (PrcB). The enzyme displayed chymotrypsin-like activity on synthetic substrates and was sensitive to peptide aldehyde and peptide vinyl sulfone inhibitors and to the Streptomyces metabolite lactacystin. Characterization of the structural genes revealed an operon-like gene organization (prcBA) similar to Rhodococcus and Mycobacterium spp. and showed that the beta subunit is encoded with a 53-amino-acid propeptide which is removed during proteasome assembly. The upstream DNA region contains the conserved orf7 and an AAA ATPase gene (arc).
Figures




References
-
- Akopian T N, Kisselev A F, Goldberg A L. Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J Biol Chem. 1997;272:1791–1798. - PubMed
-
- Baumeister W, Dahlmann B, Hegerl R, Kopp F, Kuehn L, Pfeifer G. Electron microscopy and image analysis of the multicatalytic proteinase. FEBS Lett. 1988;241:239–245. - PubMed
-
- Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

