Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments
- PMID: 9770453
- PMCID: PMC22798
- DOI: 10.1073/pnas.95.21.12141
Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments
Abstract
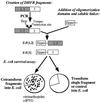
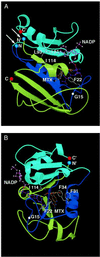
Reassembly of enzymes from peptide fragments has been used as a strategy for understanding the evolution, folding, and role of individual subdomains in catalysis and regulation of activity. We demonstrate an oligomerization-assisted enzyme reassembly strategy whereby fragments are covalently linked to independently folding and interacting domains whose interactions serve to promote efficient refolding and complementation of fragments, forming active enzyme. We show that active murine dihydrofolate reductase (E.C. 1.5.1.3) can be reassembled from complementary N- and C-terminal fragments when fused to homodimerizing GCN4 leucine zipper-forming sequences as well as heterodimerizing protein partners. Reassembly is detected by an in vivo selection assay in Escherichia coli and in vitro. The effects of mutations that disrupt fragment affinity or enzyme activity were assessed. The steady-state kinetic parameters for the reassembled mutant (Phe-31 --> Ser) were determined; they are not significantly different from the full-length mutant. The strategy described here provides a general approach for protein dissection and domain swapping studies, with the capacity both for rapid in vivo screening as well as in vitro characterization. Further, the strategy suggests a simple in vivo enzyme-based detection system for protein-protein interactions, which we illustrate with two examples: ras-GTPase and raf-ras-binding domain and FK506-binding protein-rapamycin complexed with the target of rapamycin TOR2.
Figures


References
-
- Ullmann A, Jacob F, Monod J. J Mol Biol. 1967;24:339–343. - PubMed
-
- Taniuchi H, Anfinsen C B. J Biol Chem. 1971;246:2291–2301. - PubMed
-
- Ladurner A G, Itzhaki L S, Gay G D, Fersht A R. J Mol Biol. 1997;273:317–329. - PubMed
-
- de Prat Gay G, Ruiz-Sanz J, Fersht A R. Biochemistry. 1994;33:7964–7970. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

