Activation of microhelix charging by localized helix destabilization
- PMID: 9770466
- PMCID: PMC22811
- DOI: 10.1073/pnas.95.21.12214
Activation of microhelix charging by localized helix destabilization
Abstract
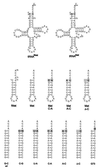
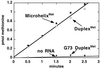
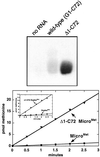
We report that aminoacylation of minimal RNA helical substrates is enhanced by mismatched or unpaired nucleotides at the first position in the helix. Previously, we demonstrated that the class I methionyl-tRNA synthetase aminoacylates RNA microhelices based on the acceptor stem of initiator and elongator tRNAs with greatly reduced efficiency relative to full-length tRNA substrates. The cocrystal structure of the class I glutaminyl-tRNA synthetase with tRNAGln revealed an uncoupling of the first (1.72) base pair of tRNAGln, and tRNAMet was proposed by others to have a similar base-pair uncoupling when bound to methionyl-tRNA synthetase. Because the anticodon is important for efficient charging of methionine tRNA, we thought that 1.72 distortion is probably effected by the synthetase-anticodon interaction. Small RNA substrates (minihelices, microhelices, and duplexes) are devoid of the anticodon triplet and may, therefore, be inefficiently aminoacylated because of a lack of anticodon-triggered acceptor stem distortion. To test this hypothesis, we constructed microhelices that vary in their ability to form a 1.72 base pair. The results of kinetic assays show that microhelix aminoacylation is activated by destabilization of this terminal base pair. The largest effect is seen when one of the two nucleotides of the pair is completely deleted. Activation of aminoacylation is also seen with the analogous deletion in a minihelix substrate for the closely related isoleucine enzyme. Thus, for at least the methionine and isoleucine systems, a built-in helix destabilization compensates in part for the lack of presumptive anticodon-induced acceptor stem distortion.
Figures


Similar articles
-
Microhelix aminoacylation by a class I tRNA synthetase. Non-conserved base pairs required for specificity.J Biol Chem. 1993 Mar 25;268(9):6069-72. J Biol Chem. 1993. PMID: 7681057
-
Evidence that specificity of microhelix charging by a class I tRNA synthetase occurs in the transition state of catalysis.Biochemistry. 1996 Jan 16;35(2):608-15. doi: 10.1021/bi9520904. Biochemistry. 1996. PMID: 8555234
-
The presence of a D-stem but not a T-stem is essential for triggering aminoacylation upon anticodon binding in yeast methionine tRNA.J Mol Biol. 1995 May 26;249(1):45-58. doi: 10.1006/jmbi.1995.0279. J Mol Biol. 1995. PMID: 7776375
-
Yeast tRNA(Met) recognition by methionyl-tRNA synthetase requires determinants from the primary, secondary and tertiary structure: a review.Biochimie. 1996;78(7):597-604. doi: 10.1016/s0300-9084(96)80006-x. Biochimie. 1996. PMID: 8955903 Review.
-
Small RNA helices as substrates for aminoacylation and their relationship to charging of transfer RNAs.Eur J Biochem. 1992 Jun 1;206(2):315-21. doi: 10.1111/j.1432-1033.1992.tb16929.x. Eur J Biochem. 1992. PMID: 1375910 Review.
Cited by
-
A recurrent general RNA binding domain appended to plant methionyl-tRNA synthetase acts as a cis-acting cofactor for aminoacylation.EMBO J. 2000 Dec 15;19(24):6908-17. doi: 10.1093/emboj/19.24.6908. EMBO J. 2000. PMID: 11118226 Free PMC article.
-
Comparison and functional implications of the 3D architectures of viral tRNA-like structures.RNA. 2009 Feb;15(2):294-307. doi: 10.1261/rna.1360709. RNA. 2009. PMID: 19144910 Free PMC article.
-
MEHMO syndrome mutation EIF2S3-I259M impairs initiator Met-tRNAiMet binding to eukaryotic translation initiation factor eIF2.Nucleic Acids Res. 2019 Jan 25;47(2):855-867. doi: 10.1093/nar/gky1213. Nucleic Acids Res. 2019. PMID: 30517694 Free PMC article.
-
Polyamine Control of Translation Elongation Regulates Start Site Selection on Antizyme Inhibitor mRNA via Ribosome Queuing.Mol Cell. 2018 Apr 19;70(2):254-264.e6. doi: 10.1016/j.molcel.2018.03.015. Mol Cell. 2018. PMID: 29677493 Free PMC article.
-
Amino acid substrates impose polyamine, eIF5A, or hypusine requirement for peptide synthesis.Nucleic Acids Res. 2017 Aug 21;45(14):8392-8402. doi: 10.1093/nar/gkx532. Nucleic Acids Res. 2017. PMID: 28637321 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

