The structure and organization within the membrane of the helices composing the pore-forming domain of Bacillus thuringiensis delta-endotoxin are consistent with an "umbrella-like" structure of the pore
- PMID: 9770479
- PMCID: PMC22824
- DOI: 10.1073/pnas.95.21.12289
The structure and organization within the membrane of the helices composing the pore-forming domain of Bacillus thuringiensis delta-endotoxin are consistent with an "umbrella-like" structure of the pore
Abstract
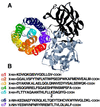
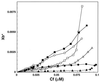
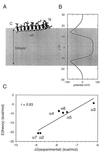
The aim of this study was to elucidate the mechanism of membrane insertion and the structural organization of pores formed by Bacillus thuringiensis delta-endotoxin. We determined the relative affinities for membranes of peptides corresponding to the seven helices that compose the toxin pore-forming domain, their modes of membrane interaction, their structures within membranes, and their orientations relative to the membrane normal. In addition, we used resonance energy transfer measurements of all possible combinatorial pairs of membrane-bound helices to map the network of interactions between helices in their membrane-bound state. The interaction of the helices with the bilayer membrane was also probed by a Monte Carlo simulation protocol to determine lowest-energy orientations. Our results are consistent with a situation in which helices alpha4 and alpha5 insert into the membrane as a helical hairpin in an antiparallel manner, while the other helices lie on the membrane surface like the ribs of an umbrella (the "umbrella model"). Our results also support the suggestion that alpha7 may serve as a binding sensor to initiate the structural rearrangement of the pore-forming domain.
Figures



References
-
- Knowles B H. Adv Insect Physiol. 1994;24:275–308.
-
- Li J D, Carroll J, Ellar D J. Nature (London) 1991;353:815–821. - PubMed
-
- Lemmon M A, Engelman D M. Q Rev Biophys. 1994;27:157–218. - PubMed
-
- Van Rie J, McGaughey W H, Johnson D E, Barnett B D, Van Mellaert H. Science. 1990;247:72–74. - PubMed
-
- Ahmad W, Ellar D J. FEMS Microbiol Lett. 1990;68:97–104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

