Linkage disequilibrium mapping in isolated populations: the example of Finland revisited
- PMID: 9770501
- PMCID: PMC22846
- DOI: 10.1073/pnas.95.21.12416
Linkage disequilibrium mapping in isolated populations: the example of Finland revisited
Abstract
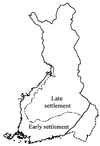
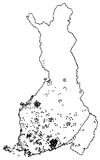
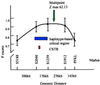
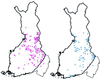
Linkage disequilibrium analysis can provide high resolution in the mapping of disease genes because it incorporates information on recombinations that have occurred during the entire period from the mutational event to the present. A circumstance particularly favorable for high-resolution mapping is when a single founding mutation segregates in an isolated population. We review here the population structure of Finland in which a small founder population some 100 generations ago has expanded into 5.1 million people today. Among the 30-odd autosomal recessive disorders that are more prevalent in Finland than elsewhere, several appear to have segregated for this entire period in the "panmictic" southern Finnish population. Linkage disequilibrium analysis has allowed precise mapping and determination of genetic distances at the 0.1-cM level in several of these disorders. Estimates of genetic distance have proven accurate, but previous calculations of the confidence intervals were too small because sampling variation was ignored. In the north and east of Finland the population can be viewed as having been "founded" only after 1500. Disease mutations that have undergone such a founding bottleneck only 20 or so generations ago exhibit linkage disequilibrium and haplotype sharing over long genetic distances (5-15 cM). These features have been successfully exploited in the mapping and cloning of many genes. We review the statistical issues of fine mapping by linkage disequilibrium and suggest that improved methodologies may be necessary to map diseases of complex etiology that may have arisen from multiple founding mutations.
Figures
References
-
- Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, et al. Nature (London) 1996;380:152–154. - PubMed
-
- Ramsay G. Nat Biotech. 1998;16:40–44. - PubMed
-
- Wang D G, Fan J-B, Siao C-J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J. Science. 1998;280:1077–1082. - PubMed
-
- Peterson A C, Rienzo A D, Lehesjoki A-E, de la Chapelle A, Slatkin M, Freimer N B. Hum Mol Genet. 1995;4:887–894. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

