Discrimination of the closely related A and D genomes of the hexaploid oat Avena sativa L
- PMID: 9770506
- PMCID: PMC22851
- DOI: 10.1073/pnas.95.21.12450
Discrimination of the closely related A and D genomes of the hexaploid oat Avena sativa L
Abstract
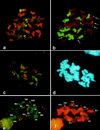
A satellite DNA sequence, As120a, specific to the A-genome chromosomes in the hexaploid oat, Avena sativa L., was isolated by subcloning a fragment with internal tandem repeats from a plasmid, pAs120, that had been obtained from an Avena strigosa (As genome) genomic library. Southern and in situ hybridization showed that sequences with homology to sequences within pAs120 were dispersed throughout the genome of diploid (A and C genomes), tetraploid (AC genomes), and hexaploid (ACD genomes) Avena species. In contrast, sequences homologous to As120a were found in two A-genome species (A. strigosa and Avena longiglumis) and in the hexaploid A. sativa whereas this sequence was little amplified in the tetraploid Avena murphyi and was absent in the remaining A- and C-genome diploid species. In situ hybridization of pAs120a to hexaploid oat species revealed the distribution of elements of the As120a repeated family over both arms of 14 of 42 chromosomes of this species. By using double in situ hybridization with pAs120a and a C genome-specific probe, three sets of 14 chromosomes were revealed corresponding to the A, C, and D genomes of the hexaploid species. Simultaneous in situ hybridizations with pAs120a and ribosomal probes were used to assign the SAT chromosomes of hexaploid species to their correct genomes. This work reports a sequence able to distinguish between the closely related A and D genomes of hexaploid oats. This sequence offers new opportunities to analyze the relationships of Avena species and to explore the possible evolution of various polyploid oat species.
Figures




References
-
- Rajhathy T, Thomas H. Cytogenetics of Oats. Ottawa: Miscellaneous Publication of the Genetics Society of Canada; 1974. , No. 2, pp. 1–90.
-
- Thomas H. In: Oat Science and Technology. Marshall H G, Sorrells M E, editors. Madison, WI: American Society of Agronomy and Crop Science Society of America; 1992. pp. 473–507.
-
- Fominaya A, Vega C, Ferrer E. Genome. 1988;30:627–632.
-
- Sánchez de la Hoz P, Fominaya A. Theor Appl Genet. 1989;77:735–741. - PubMed
-
- Linares C, Vega C, Ferrer E, Fominaya A. Theor Appl Genet. 1992;83:650–654. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

