Diversification, expression, and gamma delta T cell recognition of evolutionarily distant members of the MIC family of major histocompatibility complex class I-related molecules
- PMID: 9770516
- PMCID: PMC22861
- DOI: 10.1073/pnas.95.21.12510
Diversification, expression, and gamma delta T cell recognition of evolutionarily distant members of the MIC family of major histocompatibility complex class I-related molecules
Abstract
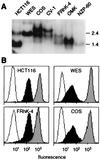
Distant relatives of major histocompatibility complex (MHC) class I molecules, human MICA and MICB, function as stress-induced antigens that are broadly recognized by intestinal epithelial gamma delta T cells. They may thus play a central role in the immune surveillance of damaged, infected, or otherwise stressed intestinal epithelial cells. However, the generality of this system in evolution and the mode of recognition of MICA and MICB are undefined. Analysis of cDNA sequences from various primate species defined translation products that are homologous to MICA and MICB. All of the MIC polypeptides have common characteristics, although they are extraordinarily diverse. The most notable alterations are several deletions and frequent amino acid substitutions in the putative alpha-helical regions of the alpha1 alpha2 domains. However, the primate MIC molecules were expressed on the surfaces of normal and transfected cells. Moreover, despite their sharing of relatively few identical amino acids in potentially accessible regions of their alpha1 alpha2 domains, they were recognized by diverse human intestinal epithelial gamma delta T cells that are restricted by MICA and MICB. Thus, MIC molecules represent a family of MHC proteins that are structurally diverse yet appear to be functionally conserved. The promiscuous mode of gamma delta T cell recognition of these antigens may be explained by their sharing of a single conserved interaction site.
Figures


Similar articles
-
Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells.Science. 1998 Mar 13;279(5357):1737-40. doi: 10.1126/science.279.5357.1737. Science. 1998. PMID: 9497295
-
Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB.Proc Natl Acad Sci U S A. 1999 Jun 8;96(12):6879-84. doi: 10.1073/pnas.96.12.6879. Proc Natl Acad Sci U S A. 1999. PMID: 10359807 Free PMC article.
-
A second lineage of mammalian major histocompatibility complex class I genes.Proc Natl Acad Sci U S A. 1994 Jul 5;91(14):6259-63. doi: 10.1073/pnas.91.14.6259. Proc Natl Acad Sci U S A. 1994. PMID: 8022771 Free PMC article.
-
Recognition of MHC TL gene products by gamma delta T cells.Immunol Rev. 1991 Apr;120:89-115. doi: 10.1111/j.1600-065x.1991.tb00589.x. Immunol Rev. 1991. PMID: 1830863 Review.
-
Structure and function of H-2 T (Tla) region class I MHC molecules.Crit Rev Immunol. 1994;14(1):1-27. Crit Rev Immunol. 1994. PMID: 7741975 Review.
Cited by
-
Immunogenetics of the NKG2D ligand gene family.Immunogenetics. 2012 Dec;64(12):855-67. doi: 10.1007/s00251-012-0638-9. Epub 2012 Jul 29. Immunogenetics. 2012. PMID: 22843249 Review.
-
Natural Killer Cell Evasion Is Essential for Infection by Rhesus Cytomegalovirus.PLoS Pathog. 2016 Aug 31;12(8):e1005868. doi: 10.1371/journal.ppat.1005868. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27580123 Free PMC article.
-
The HIV-1 pandemic: does the selective sweep in chimpanzees mirror humankind's future?Retrovirology. 2013 May 24;10:53. doi: 10.1186/1742-4690-10-53. Retrovirology. 2013. PMID: 23705941 Free PMC article. Review.
-
Pathogenesis of Adamantiades-Behçet's disease.Med Microbiol Immunol. 2003 Aug;192(3):149-55. doi: 10.1007/s00430-002-0167-5. Epub 2003 Mar 5. Med Microbiol Immunol. 2003. PMID: 12684757 Review.
-
Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence.Proc Natl Acad Sci U S A. 2003 Jun 24;100(13):7708-13. doi: 10.1073/pnas.1230533100. Epub 2003 Jun 10. Proc Natl Acad Sci U S A. 2003. PMID: 12799463 Free PMC article.
References
-
- Beckman E M, Brenner M B. Immunol Today. 1995;16:349–352. - PubMed
-
- Germain R N, Margulies D H. Annu Rev Immunol. 1993;11:403–450. - PubMed
-
- Bjorkman P J, Parham P. Annu Rev Biochem. 1990;59:253–288. - PubMed
-
- Madden D R. Annu Rev Immunol. 1995;13:587–622. - PubMed
-
- Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature (London) 1996;384:134–141. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

