Crystal structure of Taq DNA polymerase in complex with an inhibitory Fab: the Fab is directed against an intermediate in the helix-coil dynamics of the enzyme
- PMID: 9770525
- PMCID: PMC22870
- DOI: 10.1073/pnas.95.21.12562
Crystal structure of Taq DNA polymerase in complex with an inhibitory Fab: the Fab is directed against an intermediate in the helix-coil dynamics of the enzyme
Abstract
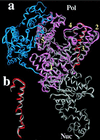
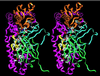
We report the crystal structure of Thermus aquaticus DNA polymerase I in complex with an inhibitory Fab, TP7, directed against the native enzyme. Some of the residues present in a helical conformation in the native enzyme have adopted a gamma turn conformation in the complex. Taken together, structural information that describes alteration of helical structure and solution studies that demonstrate the ability of TP7 to inhibit 100% of the polymerase activity of the enzyme suggest that the change in conformation is probably caused by trapping of an intermediate in the helix-coil dynamics of this helix by the Fab. Antibodies directed against modified helices in proteins have long been anticipated. The present structure provides direct crystallographic evidence. The Fab binds within the DNA binding cleft of the polymerase domain, interacting with several residues that are used by the enzyme in binding the primer:template complex. This result unequivocally corroborates inferences drawn from binding experiments and modeling calculations that the inhibitory activity of this Fab is directly attributable to its interference with DNA binding by the polymerase domain of the enzyme. The combination of interactions made by the Fab residues in both the polymerase and the vestigial editing nuclease domain of the enzyme reveal the structural basis of its preference for binding to DNA polymerases of the Thermus species. The orientation of the structure-specific nuclease domain with respect to the polymerase domain is significantly different from that seen in other structures of this polymerase. This reorientation does not appear to be antibody-induced and implies remarkably high relative mobility between these two domains.
Figures


Similar articles
-
Structure of taq DNA polymerase shows a new orientation for the structure-specific nuclease domain.Acta Crystallogr D Biol Crystallogr. 1999 Dec;55(Pt 12):1971-7. doi: 10.1107/s090744499901135x. Acta Crystallogr D Biol Crystallogr. 1999. Retraction in: Acta Crystallogr D Biol Crystallogr. 2010 Feb;66(Pt 2):222. doi: 10.1107/S0907444910000259. PMID: 10666572 Retracted.
-
Structural studies on an inhibitory antibody against Thermus aquaticus DNA polymerase suggest mode of inhibition.Protein Eng. 1998 Feb;11(2):79-86. doi: 10.1093/protein/11.2.79. Protein Eng. 1998. PMID: 9605541
-
Global conformations, hydrodynamics, and X-ray scattering properties of Taq and Escherichia coli DNA polymerases in solution.J Biol Chem. 2003 Jul 11;278(28):25341-7. doi: 10.1074/jbc.M302118200. Epub 2003 May 3. J Biol Chem. 2003. PMID: 12730189
-
Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.J Mol Recognit. 1999 Jan-Feb;12(1):19-32. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<19::AID-JMR445>3.0.CO;2-Y. J Mol Recognit. 1999. PMID: 10398393 Review.
-
[Comparative Analysis of Family A DNA-Polymerases as a Searching Tool for Enzymes with New Properties].Mol Biol (Mosk). 2023 Mar-Apr;57(2):185-196. Mol Biol (Mosk). 2023. PMID: 37000648 Review. Russian.
Cited by
-
Loop-Mediated Isothermal Amplification: From Theory to Practice.Russ J Bioorg Chem. 2022;48(6):1159-1174. doi: 10.1134/S106816202206022X. Epub 2022 Dec 23. Russ J Bioorg Chem. 2022. PMID: 36590469 Free PMC article.
-
Dynamic allostery: linkers are not merely flexible.Structure. 2011 Jul 13;19(7):907-17. doi: 10.1016/j.str.2011.06.002. Structure. 2011. PMID: 21742258 Free PMC article. Review.
-
Structure, function and evolution of the bacterial DinG-like proteins.Comput Struct Biotechnol J. 2025 Mar 17;27:1124-1139. doi: 10.1016/j.csbj.2025.03.023. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40206346 Free PMC article. Review.
-
A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro.Nucleic Acids Res. 2004 Feb 18;32(3):1197-207. doi: 10.1093/nar/gkh271. Print 2004. Nucleic Acids Res. 2004. PMID: 14973201 Free PMC article.
-
Engineering of a thermostable viral polymerase using metagenome-derived diversity for highly sensitive and specific RT-PCR.Nucleic Acids Res. 2019 Apr 23;47(7):3619-3630. doi: 10.1093/nar/gkz104. Nucleic Acids Res. 2019. PMID: 30767012 Free PMC article.
References
-
- Sharkey D J, Scalice E R, Christy K G, Atwood S M, Daiss J L. Bio/Technology. 1994;12:506–509. - PubMed
-
- Scalice E R, Sharkey D J, Daiss J L. J Immunol Methods. 1995;183:15–26. - PubMed
-
- Murali R, Helmer-Citterich M, Sharkey D J, Scalice E R, Daiss J L, Sullivan M A, Krishna Murthy H M. Protein Eng. 1998;11:79–86. - PubMed
-
- Kim Y, Eom S H, Wang J, Lee D, Suh S, Steitz T A. Nature (London) 1995;376:612–616. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

