Position-dependent methylation and transcriptional silencing of transgenes in inverted T-DNA repeats: implications for posttranscriptional silencing of homologous host genes in plants
- PMID: 9774634
- PMCID: PMC109204
- DOI: 10.1128/MCB.18.11.6165
Position-dependent methylation and transcriptional silencing of transgenes in inverted T-DNA repeats: implications for posttranscriptional silencing of homologous host genes in plants
Abstract
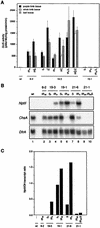
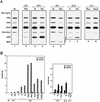
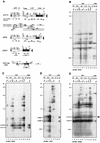
Posttranscriptional silencing of chalcone synthase (Chs) genes in petunia transformants occurs by introducing T-DNAs that contain a promoter-driven or promoterless Chs transgene. With the constructs we used, silencing occurs only by T-DNA loci which are composed of two or more T-DNA copies that are arranged as inverted repeats (IRs). Since we are interested in the mechanism by which these IR loci induce silencing, we have analyzed different IR loci and nonsilencing single-copy (S) T-DNA loci with respect to the expression and methylation of the transgenes residing in these loci. We show that in an IR locus, the transgenes located proximal to the IR center are much more highly methylated than are the distal genes. A strong silencing locus composed of three inverted T-DNAs bearing promoterless Chs transgenes was methylated across the entire locus. The host Chs genes in untransformed plants were moderately methylated, and no change in methylation was detected when the genes were silenced. Run-on transcription assays showed that promoter-driven transgenes located proximal to the center of a particular IR are transcriptionally more repressed than are the distal genes of the same IR locus. Transcription of the promoterless Chs transgenes could not be detected. In the primary transformant, some of the IR loci were detected together with an unlinked S locus. We observed that the methylation and expression characteristics of the transgenes of these S loci were comparable to those of the partner IR loci, suggesting that there has been cross talk between the two types of loci. Despite the similar features, S loci are unable to induce silencing, indicating that the palindromic arrangement of the Chs transgenes in the IR loci is critical for silencing. Since transcriptionally silenced transgenes in IRs can trigger posttranscriptional silencing of the host genes, our data are most consistent with a model of silencing in which the transgenes physically interact with the homologous host gene(s). The interaction may alter epigenetic features other than methylation, thereby impairing the regular production of mRNA.
Figures



References
-
- Assaad F F, Tucker K L, Signer E R. Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol Biol. 1993;22:1067–1085. - PubMed
-
- Barlow D P. Gametic imprinting in mammals. Science. 1995;270:1610–1613. - PubMed
-
- Baulcombe D C, English J J. Ectopic pairing of homologous DNA and post-transcriptional silencing in transgenic plants. Curr Opin Biotechnol. 1996;7:173–180.
-
- Bender J, Fink G R. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell. 1995;83:725–734. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
