Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA
- PMID: 9774635
- PMCID: PMC109205
- DOI: 10.1128/MCB.18.11.6178
Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA
Abstract
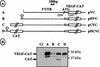
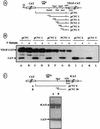
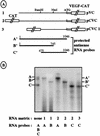
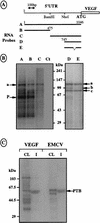
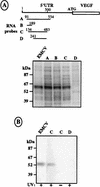
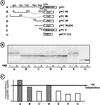
The mRNA of vascular endothelial growth factor (VEGF), the major angiogenic growth factor, contains an unusually long (1,038 nucleotides) and structured 5' untranslated region (UTR). According to the classical translation initiation model of ribosome scanning, such a 5' UTR is expected to be a strong translation inhibitor. In vitro and bicistronic strategies were used to show that the VEGF mRNA translation was cap independent and occurred by an internal ribosome entry process. For the first time, we demonstrate that two independent internal ribosome entry sites (IRESs) are present in this 5' UTR. IRES A is located within the 300 nucleotides upstream from the AUG start codon. RNA secondary structure prediction and site-directed mutagenesis allowed the identification of a 49-nucleotide structural domain (D4) essential to IRES A activity. UV cross-linking experiments revealed that IRES A activity was correlated with binding of a 100-kDa protein to the D4 domain. IRES B is located in the first half of the 5' UTR. An element between nucleotides 379 and 483 is required for its activity. Immunoprecipitation experiments demonstrated that a main IRES B-bound protein was the polypyrimidine tract binding protein (PTB), a well-known regulator of picornavirus IRESs. However, we showed that binding of the PTB on IRES B does not seem to be correlated with its activity. Evidence is provided of an original cumulative effect of two IRESs, probably controlled by different factors, to promote an efficient initiation of translation at the same AUG codon.
Figures


References
-
- Bernstein J, Sella O, Le S Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. - PubMed
-
- Borman A, Howell M T, Patton J G, Jackson R J. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J Gen Virol. 1993;74:1775–1788. - PubMed
-
- Borovjagin A, Pestova T, Shatsky I. Pyrimidine tract binding protein strongly stimulates in vitro encephalomyocarditis virus RNA translation at the level of preinitiation complex formation. FEBS Lett. 1994;351:299–302. - PubMed
-
- Brogi E, Wu T, Namiki A, Isner J M. Indirect angiogenic cytokines upregulate VEGF and bFGF gene expression in vascular smooth muscle cells, whereas hypoxia upregulates VEGF expression only. Circulation. 1994;90:649–652. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
