MFR, a putative receptor mediating the fusion of macrophages
- PMID: 9774638
- PMCID: PMC109208
- DOI: 10.1128/MCB.18.11.6213
MFR, a putative receptor mediating the fusion of macrophages
Abstract
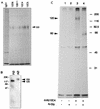
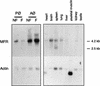
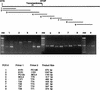
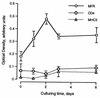
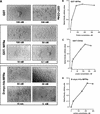
We had previously identified a macrophage surface protein whose expression is highly induced, transient, and specific, as it is restricted to actively fusing macrophages in vitro and in vivo. This protein is recognized by monoclonal antibodies that block macrophage fusion. We have now purified this protein and cloned its corresponding cDNA. This protein belongs to the superfamily of immunoglobulins and is similar to immune antigen receptors such as the T-cell receptor, B-cell receptor, and viral receptors such as CD4. We have therefore named this protein macrophage fusion receptor (MFR). We show that the extracellular domain of MFR prevents fusion of macrophages in vitro and therefore propose that MFR belongs to the fusion machinery of macrophages. MFR is identical to SHPS-1 and BIT and is a homologue of P84, SIRPalpha, and MyD-1, all of which have been recently cloned and implicated in cell signaling and cell-cell interaction events.
Figures


References
-
- Alkhatib G, Combadiere C, Broder C, Feng Y, Kennedy P, Murphy P, Berger E. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. - PubMed
-
- Bergelson J, Cunningham J, Droguett G, Kurt-Jones E, Krithivas A, Hong J, Horwitz M, Crowell R, Finberg R. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. - PubMed
-
- Blobel C, Wolfsberg T, Turck C, Myles D, Primakoff P, White J. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature. 1992;356:248–252. - PubMed
-
- Brooke G, Parsons K, Howard C. Cloning of two members of the SIRPα family of protein tyrosine phosphatase binding proteins in cattle that are expressed on monocytes and a subpopulation of dendritic cells and which mediate binding to CD4 T cells. Eur J Immunol. 1998;28:1–11. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
