Evidence that immunoglobulin VH-DJ recombination does not require germ line transcription of the recombining variable gene segment
- PMID: 9774642
- PMCID: PMC109212
- DOI: 10.1128/MCB.18.11.6253
Evidence that immunoglobulin VH-DJ recombination does not require germ line transcription of the recombining variable gene segment
Abstract
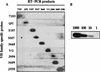
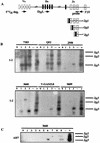
The importance of V(D)J recombination for generating diversity in the immune system is well established, but the mechanisms which regulate V(D)J recombination are still poorly understood. Although transcription of unrearranged (germ line) immunoglobulin and T-cell receptor gene segments often precedes V(D)J recombination and has been implicated in its control, the actual role of germ line transcripts in V(D)J recombination is not known. We used a sensitive reverse transcription-PCR assay to study immunoglobulin VH germ line transcripts in proB lines from RAG-deficient mice. All 10 VH families analyzed were germ line transcribed, and germ line transcription was found in all of the cell lines examined, indicating that active chromatin was present in the VH region. However, not all VH families were germ line transcribed in every cell line, and there was a surprising lack of uniformity in the number and family distribution of germ line VH transcripts in individual lines. When V(D)J recombination was activated by restoration of RAG activity, recombinational activity of endogenous VH genes for which germ line transcription was observed could be compared with those of genes for which it was not observed. This analysis revealed multiple examples of endogenous VH gene segments which were rearranged in cells where their germ line transcription was not detectable prior to RAG expression. Thus, our data provide strong support for the idea that V-(D)J recombination does not require germ line transcription of the recombining variable gene segment.
Figures





References
-
- Alt F W, Oltz E M, Young F, Gorman J, Taccioli G, Chen J. VDJ recombination. Immunol Today. 1992;13:306–314. - PubMed
-
- Alt F W, Blackwell T, Yancopoulos G. Development of the primary antibody repertoire. Science. 1987;238:1079–1087. - PubMed
-
- Alvarez J D, Anderson S J, Loh D Y. V(D)J recombination and allelic exclusion of a TCR beta-chain minilocus occurs in the absence of a functional promoter. J Immunol. 1995;155:1191–1202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
