Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA
- PMID: 9774653
- PMCID: PMC109223
- DOI: 10.1128/MCB.18.11.6374
Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA
Abstract
Saccharomyces cerevisiae Los1p, which is genetically linked to the nuclear pore protein Nsp1p and several tRNA biogenesis factors, was recently grouped into the family of importin/karyopherin-beta-like proteins on the basis of its sequence similarity. In a two-hybrid screen, we identified Nup2p as a nucleoporin interacting with Los1p. Subsequent purification of Los1p from yeast demonstrates its physical association not only with Nup2p but also with Nsp1p. By the use of the Gsp1p-G21V mutant, Los1p was shown to preferentially bind to the GTP-bound form of yeast Ran. Furthermore, overexpression of full-length or N-terminally truncated Los1p was shown to have dominant-negative effects on cell growth and different nuclear export pathways. Finally, Los1p could interact with Gsp1p-GTP, but only in the presence of tRNA, as revealed in an indirect in vitro binding assay. These data confirm the homology between Los1p and the recently identified human exportin for tRNA and reinforce the possibility of a role for Los1p in nuclear export of tRNA in yeast.
Figures
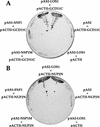
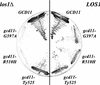


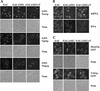

References
-
- Aitchison J D, Blobel G, Rout M P. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. - PubMed
-
- Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. - PubMed
-
- Arts G-J, Fornerod M, Mattaj I W. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. - PubMed
-
- Bailer, S. Unpublished data.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
