Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52
- PMID: 9774659
- PMCID: PMC109229
- DOI: 10.1128/MCB.18.11.6430
Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52
Abstract
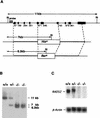
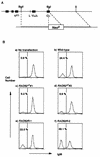
Rad52 plays a pivotal role in double-strand break (DSB) repair and genetic recombination in Saccharomyces cerevisiae, where mutation of this gene leads to extreme X-ray sensitivity and defective recombination. Yeast Rad51 and Rad52 interact, as do their human homologues, which stimulates Rad51-mediated DNA strand exchange in vitro, suggesting that Rad51 and Rad52 act cooperatively. To define the role of Rad52 in vertebrates, we generated RAD52(-/-) mutants of the chicken B-cell line DT40. Surprisingly, RAD52(-/-) cells were not hypersensitive to DNA damages induced by gamma-irradiation, methyl methanesulfonate, or cis-platinum(II)diammine dichloride (cisplatin). Intrachromosomal recombination, measured by immunoglobulin gene conversion, and radiation-induced Rad51 nuclear focus formation, which is a putative intermediate step during recombinational repair, occurred as frequently in RAD52(-/-) cells as in wild-type cells. Targeted integration frequencies, however, were consistently reduced in RAD52(-/-) cells, showing a clear role for Rad52 in genetic recombination. These findings reveal striking differences between S. cerevisiae and vertebrates in the functions of RAD51 and RAD52.
Figures


References
-
- Bai Y, Symington L S. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. - PubMed
-
- Baumann P, Benson F E, West S C. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. - PubMed
-
- Benson F E, Baumann P, West S C. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. - PubMed
-
- Bezzubova O Y, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde J-M. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell. 1997;89:185–193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
