DNA supercoiling factor localizes to puffs on polytene chromosomes in Drosophila melanogaster
- PMID: 9774687
- PMCID: PMC109257
- DOI: 10.1128/MCB.18.11.6737
DNA supercoiling factor localizes to puffs on polytene chromosomes in Drosophila melanogaster
Abstract
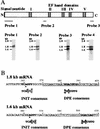
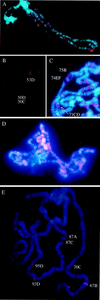
DNA supercoiling factor (SCF) was first identified in silkworm as a protein that generates negative supercoils in DNA in conjunction with eukaryotic topoisomerase II. To analyze the in vivo role of the factor, we cloned a cDNA encoding Drosophila melanogaster SCF. Northern analysis revealed 1.6- and 1.8-kb mRNAs throughout development. The longer mRNA contains an open reading frame that shares homology with mouse reticulocalbin whereas the shorter one encodes a truncated version lacking the N-terminal signal peptide-like sequence. An antibody against SCF detected a 45-kDa protein in the cytoplasmic fraction and a 30-kDa protein in the nuclear fraction of embryonic extracts. Immunoprecipitation suggests that the 30-kDa protein interacts with topoisomerase II in the nucleus, and hence that it is a functional form of SCF. Immunostaining of blastoderm embryos showed that SCF is present in nuclei during interphase but is excluded from mitotic chromosomes. In larvae, the antibody stained the nuclei of several tissues including a posterior part of the salivary gland. This latter staining was associated with natural or ecdysteroid-induced puffs on polytene chromosomes. Upon heat treatment of larvae, the staining on the endogenous puffs disappeared, and strong staining appeared on heat shock puffs. These results implicate SCF in gene expression.
Figures






Similar articles
-
DNA supercoiling factor positively regulates expression of the homeotic gene Abdominal-B in Drosophila melanogaster.Genes Cells. 2007 Dec;12(12):1347-55. doi: 10.1111/j.1365-2443.2007.01140.x. Genes Cells. 2007. PMID: 18076572
-
Functional dissection of DNA supercoiling factor: EF-hand domains and C-terminal HDEF motif are essential for its activity.Genes Cells. 1999 Jan;4(1):33-40. doi: 10.1046/j.1365-2443.1999.00236.x. Genes Cells. 1999. PMID: 10231391
-
Association of SARFH (sarcoma-associated RNA-binding fly homolog) with regions of chromatin transcribed by RNA polymerase II.Mol Cell Biol. 1995 Aug;15(8):4562-71. doi: 10.1128/MCB.15.8.4562. Mol Cell Biol. 1995. PMID: 7623847 Free PMC article.
-
Cloning of a cDNA for DNA supercoiling factor reveals a distinctive Ca(2+)-binding protein.J Biol Chem. 1995 Jun 30;270(26):15571-5. doi: 10.1074/jbc.270.26.15571. J Biol Chem. 1995. PMID: 7797553
-
Expanding the Mot1 subfamily: 89B helicase encodes a new Drosophila melanogaster SNF2-related protein which binds to multiple sites on polytene chromosomes.Nucleic Acids Res. 1996 Aug 15;24(16):3121-8. doi: 10.1093/nar/24.16.3121. Nucleic Acids Res. 1996. PMID: 8774890 Free PMC article.
Cited by
-
BmCREC is an endoplasmic reticulum (ER) resident protein and required for ER/Golgi morphology.J Biol Chem. 2013 Sep 13;288(37):26649-57. doi: 10.1074/jbc.M113.463018. Epub 2013 Aug 6. J Biol Chem. 2013. PMID: 23921381 Free PMC article.
-
Soluble HLA-DQ2 expressed in S2 cells copurifies with a high affinity insect cell derived protein.Immunogenetics. 2009 Feb;61(2):81-9. doi: 10.1007/s00251-008-0338-7. Epub 2008 Nov 6. Immunogenetics. 2009. PMID: 18987854
-
Flavonoids and Melanins: a common strategy across two kingdoms.Int J Biol Sci. 2014 Oct 29;10(10):1159-70. doi: 10.7150/ijbs.9672. eCollection 2014. Int J Biol Sci. 2014. PMID: 25516714 Free PMC article. Review.
-
Calumenin-15 facilitates filopodia formation by promoting TGF-β superfamily cytokine GDF-15 transcription.Cell Death Dis. 2013 Oct 17;4(10):e870. doi: 10.1038/cddis.2013.403. Cell Death Dis. 2013. PMID: 24136234 Free PMC article.
-
Regulation of the catalytic function of topoisomerase II alpha through association with RNA.Nucleic Acids Res. 2008 Nov;36(19):6080-90. doi: 10.1093/nar/gkn614. Epub 2008 Sep 27. Nucleic Acids Res. 2008. PMID: 18820297 Free PMC article.
References
-
- Andrew D J, Scott M T. Immunological methods for mapping protein distribution on polytene chromosomes. Methods Cell Biol. 1994;14:353–370. - PubMed
-
- Androphy E J, Schiller J T, Lowy D R. Identification of the protein encoded by the E6 transforming gene of bovine papillomavirus. Science. 1985;230:442–445. - PubMed
-
- Ashburner M. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. VI. Induction by ecdysone in salivary glands of Drosophila melanogaster cultured in vitro. Chromosoma. 1972;38:255–281. - PubMed
-
- Bhat M A, Philp A V, Glover D M, Bellen H J. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell. 1996;87:1103–1114. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
