Cytoplasmic dynein intermediate-chain isoforms with different targeting properties created by tissue-specific alternative splicing
- PMID: 9774695
- PMCID: PMC109265
- DOI: 10.1128/MCB.18.11.6816
Cytoplasmic dynein intermediate-chain isoforms with different targeting properties created by tissue-specific alternative splicing
Abstract
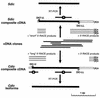
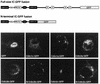
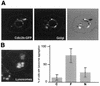
The intermediate chains (ICs) are the subunits of the cytoplasmic dynein that provide binding of the complex to cargo organelles through interaction of their N termini with dynactin. We present evidence that in Drosophila, the IC subunits are represented by at least 10 structural isoforms, created by the alternative splicing of transcripts from a unique Cdic gene. The splicing pattern is tissue specific. A constitutive set of four IC isoforms is expressed in all tissues tested; in addition, tissue-specific isoforms are found in the ovaries and nervous tissue. The structural variations between isoforms are limited to the N terminus of the IC molecule, where the interaction with dynactin takes place. This suggests differences in the dynactin-mediated organelle binding by IC isoforms. Accordingly, when transiently expressed in Drosophila Schneider-3 cells, the IC isoforms differ in their intracellular targeting properties from each other. A mechanism is proposed for the regulation of dynein binding to organelles through the changes in the content of the IC isoform pool.
Figures

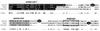




References
-
- Benevolenskaya, E. Unpublished data.
-
- Benevolenskaya E V, Nurminsky D I, Gvozdev V A. Structure of the Drosophila melanogaster annexin X gene. DNA Cell Biol. 1994;14:349–357. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
