Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores
- PMID: 9774696
- PMCID: PMC109266
- DOI: 10.1128/MCB.18.11.6826
Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores
Abstract
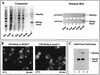
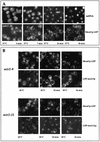
We have identified between Mex67p and Mtr2p a complex which is essential for mRNA export. This complex, either isolated from yeast or assembled in Escherichia coli, can bind in vitro to RNA through Mex67p. In vivo, Mex67p requires Mtr2p for association with the nuclear pores, which can be abolished by mutating either MEX67 or MTR2. In all cases, detachment of Mex67p from the pores into the cytoplasm correlates with a strong inhibition of mRNA export. At the nuclear pores, Nup85p represents one of the targets with which the Mex67p-Mtr2p complex interacts. Thus, Mex67p and Mtr2p constitute a novel mRNA export complex which can bind to RNA via Mex67p and which interacts with nuclear pores via Mtr2p.
Figures





References
-
- Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. - PubMed
-
- Doye V, Hurt E C. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. - PubMed
-
- Fahrenkrog, B., E. Hurt, U. Aebi, and N. Panté. Submitted for publication.
-
- Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. - PubMed
-
- Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
