Accumulated clonal genetic alterations in familial and sporadic colorectal carcinomas with widespread instability in microsatellite sequences
- PMID: 9777938
- PMCID: PMC1853059
- DOI: 10.1016/S0002-9440(10)65651-9
Accumulated clonal genetic alterations in familial and sporadic colorectal carcinomas with widespread instability in microsatellite sequences
Abstract
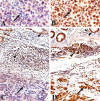
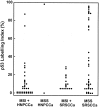
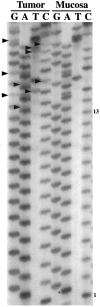
A subset of hereditary and sporadic colorectal carcinomas is defined by microsatellite instability (MSI), but the spectra of gene mutations have not been characterized extensively. Thirty-nine hereditary nonpolyposis colorectal cancer syndrome carcinomas (HNPCCa) and 57 sporadic right-sided colonic carcinomas (SRSCCa) were evaluated. Of HNPCCa, 95% (37/39) were MSI-positive as contrasted with 31% (18/57) of SRSCCa (P < 0.000001), but instability tended to be more widespread in SRSCCa (P = 0.08). Absence of nuclear hMSH2 mismatch repair gene product by immunohistochemistry was associated with germline hMSH2 mutation (P = 0.0007). The prevalence of K-ras proto-oncogene mutations was similar in HNPCCa and SRSCCa (30% (11/37) and 30% (16/54)), but no HNPCCa from patients with germline hMSH2 mutation had codon 13 mutation (P = 0.02), and two other HNPCCa had multiple K-ras mutations attributable to subclones. 18q allelic deletion and p53 gene product overexpression were inversely related to MSI (P = 0.0004 and P = 0.0001, respectively). Frameshift mutation of the transforming growth factor beta type II receptor gene was frequent in all MSI-positive cancers (85%, 46/54), but mutation of the E2F-4 transcription factor gene was more common in HNPCCa of patients with germline hMSH2 mutation than in those with germline bMLH1 mutation (100% (8/8) versus 40% (2/5), P = 0.04), and mutation of the Bax proapoptotic gene was more frequent in HNPCCa than in MSI-positive SRSCCa (55% (17/31) versus 13% (2/15), P = 0.01). The most common combination of mutations occurred in only 23% (8/35) of evaluable MSI-positive cancers. Our findings suggest that the accumulation of specific genetic alterations in MSI-positive colorectal cancers is markedly heterogeneous, because the occurrence of some mutations (eg, ras, E2F-4, and Bax genes), but not others (eg, transforming growth factor beta type II receptor gene), depends on the underlying basis of the mismatch repair deficiency. This genetic heterogeneity may contribute to the heterogeneous clinical and pathological features of MSI-positive cancers.
Figures







References
-
- Kinzler KW, Vogelstein B: Lessons from hereditary colorectal cancer. Cell 1996, 87:159-170 - PubMed
-
- Tomlinson I, Ilyas M, Novelli M: Molecular genetics of colon cancer. Cancer Metastasis Rev 1997, 16:67-79 - PubMed
-
- Gryfe R, Swallow C, Bapat B, Redston M, Gallinger S, Couture J: Molecular biology of colorectal cancer. Curr Probl Cancer 1997, 21:233-300 - PubMed
-
- Hartsough MT, Mulder KM: Transforming growth factor-β signaling in epithelial cells. Pharmacol Ther 1997, 75:21-41 - PubMed
-
- Heldin CH, Miyazono K, ten Dijke P: TGFβ signalling from cell membrane to nucleus through SMAD proteins. Nature 1997, 390:465-471 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

