Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis
- PMID: 9784541
- PMCID: PMC108667
- DOI: 10.1128/IAI.66.11.5337-5343.1998
Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis
Abstract
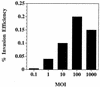
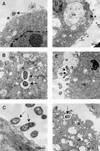
Invasion of host cells is believed to be an important strategy utilized by a number of pathogens, which affords them protection from the host immune system. The connective tissues of the periodontium are extremely well vascularized, which allows invading microorganisms, such as the periodontal pathogen Porphyromonas gingivalis, to readily enter the bloodstream. However, the ability of P. gingivalis to actively invade endothelial cells has not been previously examined. In this study, we demonstrate that P. gingivalis can invade bovine and human endothelial cells as assessed by an antibiotic protection assay and by transmission and scanning electron microscopy. P. gingivalis A7436 was demonstrated to adhere to and to invade fetal bovine heart endothelial cells (FBHEC), bovine aortic endothelial cells (BAEC), and human umbilical vein endothelial cells (HUVEC). Invasion efficiencies of 0.1, 0.2, and 0. 3% were obtained with BAEC, HUVEC, and FBHEC, respectively. Invasion of FBHEC and BAEC by P. gingivalis A7436 assessed by electron microscopy revealed the formation of microvillus-like extensions around adherent bacteria followed by the engulfment of the pathogen within vacuoles. Invasion of BAEC by P. gingivalis A7436 was inhibited by cytochalasin D, nocodazole, staurosporine, protease inhibitors, and sodium azide, indicating that cytoskeletal rearrangements, protein phosphorylation, energy metabolism, and P. gingivalis proteases are essential for invasion. In contrast, addition of rifampin, nalidixic acid, and chloramphenicol had little effect on invasion, indicating that bacterial RNA, DNA, and de novo protein synthesis are not required for P. gingivalis invasion of endothelial cells. Likewise de novo protein synthesis by endothelial cells was not required for invasion by P. gingivalis. P. gingivalis 381 was demonstrated to adhere to and to invade BAEC (0.11 and 0.1% efficiency, respectively). However, adherence and invasion of the corresponding fimA mutant DPG3, which lacks the major fimbriae, was not detected. These results indicate that P. gingivalis can actively invade endothelial cells and that fimbriae are required for this process. P. gingivalis invasion of endothelial cells may represent another strategy utilized by this pathogen to thwart the host immune response.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

